 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | SMil_00021663-RA_Salv | ||||||||
| Common Name | SPL10 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Lamiaceae; Nepetoideae; Mentheae; Salvia
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 858aa MW: 95048.2 Da PI: 6.8039 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 129.8 | 1e-40 | 10 | 87 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+Cqve+C adls+ak+yhrrhkvC++hska+++lv +++qrfCqqCsrfh+l efDe+krsCrrrLa+hn+rrrk+q+
SMil_00021663-RA_Salv 10 VCQVEDCGADLSKAKDYHRRHKVCQMHSKASKALVGNQMQRFCQQCSRFHALLEFDEGKRSCRRRLAGHNKRRRKTQT 87
6**************************************************************************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 9.7E-33 | 4 | 72 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 31.71 | 8 | 85 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 1.12E-37 | 9 | 91 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 2.0E-29 | 11 | 84 | IPR004333 | Transcription factor, SBP-box |
| Gene3D | G3DSA:1.25.40.20 | 2.7E-5 | 163 | 192 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 1.38E-5 | 638 | 749 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 9.82E-5 | 639 | 749 | No hit | No description |
| Gene3D | G3DSA:1.25.40.20 | 2.7E-5 | 642 | 752 | IPR020683 | Ankyrin repeat-containing domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0016021 | Cellular Component | integral component of membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 858 aa Download sequence Send to blast |
LAGASSNRAV CQVEDCGADL SKAKDYHRRH KVCQMHSKAS KALVGNQMQR FCQQCSRFHA 60 LLEFDEGKRS CRRRLAGHNK RRRKTQTDNM SNNSTVDNNQ TSNYLLMSIL KILSSMHSNR 120 SNHRDDQDLL SHLLQGLASQ GSLHWERNIS AHLQESPDLL NNLASVGNSE LVSMLISNGS 180 PGQTSRQEHC IALGDEMPQK IGNLHSTSQR PGIVLHTQAG SQYAQGLETS GGRSKLNNFD 240 LNDIYVDSDD GTDDMERSTA PQGLGAVSIG CPSWVQQESH QSSPPQTSGN SDSASAQSPS 300 SSSGEAQSHT DRIVFKLFGK EPSDFPIVLR SQIFDWLSHS PSDIESYIRP GCVILTIYLR 360 LSESAWEELY CDLSSSISRL LNFSDDVNFW STGWIYARVQ NQMAFVHNGQ VVVNTSLPLG 420 TDSFSSILSI KPIAVASSGK AQFIVKGVNL SRPSSRLLCA LEGNYLVANS ESMEHVDSLE 480 HVQCLKFSCS IPAVTGRGFI EVGYSTHCKV EDHGLSSSYY PFIIAEENVC AEIRMLEKEI 540 ELIEEDRLQR GTGRIAARSQ AMEFIHEMGW LLHKYQLMSR SENEESNLDF FPFERFKCLV 600 EFSMDRDWCS VVNKLLEILF SGTVSSGEKP FLKFAMSEMS LLHRAVRKNS RPLVGMLLRY 660 VPEKIADELS SEYGSLVSGY LFRPDVAGPG GLTPLHVAAG RDGSEDILDA LTDDPGKVGI 720 DAWKSALDST GFTPENYARL RGHYSYIHLV QRKMNKKAPS GHVVVDISDT LSESSINQKT 780 NAATFEIAAR SQTKPCGVCA QNLAYRSGSR TLLYRPTMLS MLAIAAVCVC VALLFKSSPR 840 VLFVFRPFSW EMLEYGSS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 5e-29 | 4 | 84 | 4 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
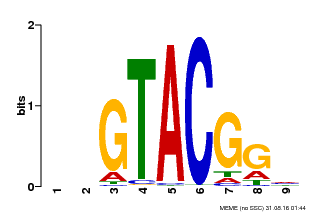
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3' of AP1 promoter. Binds specifically to the 5'-GTAC-3' core sequence. {ECO:0000269|PubMed:10524240, ECO:0000269|PubMed:16095614}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00633 | PBM | Transfer from PK06791.1 | Download |
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KF437886 | 0.0 | KF437886.1 Salvia miltiorrhiza SQUAMOSA promoter binding protein-like 10 (SPL10) mRNA, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011079486.1 | 0.0 | squamosa promoter-binding-like protein 1 | ||||
| Swissprot | Q9SMX9 | 0.0 | SPL1_ARATH; Squamosa promoter-binding-like protein 1 | ||||
| TrEMBL | A0A075FJ61 | 0.0 | A0A075FJ61_SALMI; SQUAMOSA promoter binding protein-like 10 (Fragment) | ||||
| STRING | VIT_07s0005g02260.t01 | 0.0 | (Vitis vinifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA1871 | 24 | 66 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G47070.1 | 0.0 | squamosa promoter binding protein-like 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



