 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | SMil_00021680-RA_Salv | ||||||||
| Common Name | WRI10 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Lamiaceae; Nepetoideae; Mentheae; Salvia
|
||||||||
| Family | AP2 | ||||||||
| Protein Properties | Length: 417aa MW: 45842.6 Da PI: 9.8505 | ||||||||
| Description | AP2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 53.8 | 4.9e-17 | 104 | 153 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkr.krfslgkfgtaeeAakaaiaarkkleg 55
s+y+GV++++++grW+++I+d + k+++lg f+ta Aa+a+++a+ k++g
SMil_00021680-RA_Salv 104 SQYRGVTFYRRTGRWESHIWD------CgKQVYLGGFDTAHAAARAYDRAAIKFRG 153
78*******************......55************************997 PP
| |||||||
| 2 | AP2 | 39.7 | 1.2e-12 | 196 | 243 | 1 | 52 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkr.krfslgkfgtaeeAakaaiaarkk 52
s+y+GV+ +k grW+A+ + + k+ +lg fg++ eAa+a+++a+ k
SMil_00021680-RA_Salv 196 SKYRGVTLHK-CGRWEARMGQF----IGkKYMYLGLFGSEVEAARAYDKAAIK 243
89********.7******6653....235**********99*********987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF54171 | 1.24E-15 | 104 | 161 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 4.5E-9 | 104 | 153 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 7.8E-17 | 105 | 161 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 7.5E-33 | 105 | 167 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 2.57E-11 | 105 | 160 | No hit | No description |
| PROSITE profile | PS51032 | 17.82 | 105 | 161 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 1.18E-15 | 196 | 256 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 2.5E-8 | 196 | 243 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 2.35E-23 | 196 | 256 | No hit | No description |
| Gene3D | G3DSA:3.30.730.10 | 2.2E-14 | 197 | 255 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 2.1E-28 | 197 | 260 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 15.092 | 197 | 254 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 6.1E-6 | 198 | 209 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 6.1E-6 | 236 | 256 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 417 aa Download sequence Send to blast |
MKKSVSTLNF SILDKHVIEI EDDANNNVNS SGNELQLFPI GTSPPSPTPA MVRAKYWLNL 60 SVPEVSGGGS GVELGIYKAQ LPANAPVQPP QAKKSRRGPR SRSSQYRGVT FYRRTGRWES 120 HIWDCGKQVY LGGFDTAHAA ARAYDRAAIK FRGVDADINF TIGDYDEEMK QMKSLTKEEF 180 VHILRRQSNG FARGSSKYRG VTLHKCGRWE ARMGQFIGKK YMYLGLFGSE VEAARAYDKA 240 AIKSNGREAI TNFELSNYDR EMNFNSGNGG NGSSLDLNLG ISLSSDGPQG NDTTRNPHFP 300 IQPSEFPDGK RLKVEPISAP QELMQKYPPM WAGIYAGFAP NSKELATRMS GEAVPLPGYS 360 SNWQWKMPSH GMVTPVPVMA SSAASSGFSP ATSPYFSSVV PLTNRAQFPA TAFHSLQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor (By similarity). Involved in spikelet transition (Probable). Together with SNB, controls synergistically inflorescence architecture and floral meristem establishment via the regulation of spatio-temporal expression of B- and E-function floral organ identity genes in the lodicules and of spikelet meristem genes (PubMed:22003982). Prevents lemma and palea elongation as well as grain growth (PubMed:28066457). {ECO:0000250|UniProtKB:P47927, ECO:0000269|PubMed:22003982, ECO:0000269|PubMed:28066457, ECO:0000305|PubMed:26631749}. | |||||
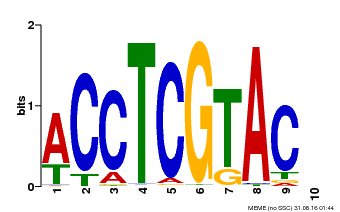
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00615 | PBM | Transfer from AT2G28550 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Target of miR172 microRNA mediated cleavage, particularly during floral organ development. {ECO:0000269|PubMed:22003982, ECO:0000305|PubMed:20017947, ECO:0000305|PubMed:26631749, ECO:0000305|PubMed:28066457}. | |||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KJ784461 | 0.0 | KJ784461.1 Salvia miltiorrhiza AP10/EREBP Transcription Factors (WRI10) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020548798.1 | 0.0 | AP2-like ethylene-responsive transcription factor TOE3 isoform X1 | ||||
| Refseq | XP_020548799.1 | 0.0 | AP2-like ethylene-responsive transcription factor TOE3 isoform X1 | ||||
| Swissprot | Q84TB5 | 2e-98 | AP22_ORYSJ; APETALA2-like protein 2 | ||||
| TrEMBL | A0A0G2SJH4 | 0.0 | A0A0G2SJH4_SALMI; AP10/EREBP Transcription Factors | ||||
| STRING | Migut.F02018.1.p | 0.0 | (Erythranthe guttata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA591 | 24 | 116 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G28550.3 | 5e-94 | related to AP2.7 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



