 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | SMil_00025248-RA_Salv | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Lamiaceae; Nepetoideae; Mentheae; Salvia
|
||||||||
| Family | Trihelix | ||||||||
| Protein Properties | Length: 437aa MW: 49267.8 Da PI: 7.8423 | ||||||||
| Description | Trihelix family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | trihelix | 38.2 | 3.8e-12 | 66 | 120 | 2 | 61 |
trihelix 2 WtkqevlaLiearremeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkry 61
W+++e laL+++r+ me ++ + We+vs+k+ e gf+rs ++Ckek+e+ + r+
SMil_00025248-RA_Salv 66 WSHDELLALLKLRSTMEIWFPDF-----TWEHVSRKLGELGFKRSGEECKEKFEEESCRH 120
********************998.....9*************************977665 PP
| |||||||
| 2 | trihelix | 93.2 | 2.6e-29 | 330 | 417 | 1 | 85 |
trihelix 1 rWtkqevlaLiearremeerlr....rgklkkplWeevskkmrergferspkqCkekwenlnkrykkikegekkrtsessstcpyfdq 84
rW+++ev+aLi++r + + + ++ ++plWe++sk m+e g++rs+k+Ckekwen+nk+++k+++++kkr s +s+tcpyf+q
SMil_00025248-RA_Salv 330 RWPRDEVQALIKLRCKAGGGGGgggeEEGGRGPLWERISKGMMEMGYKRSAKRCKEKWENINKYFRKTRDSSKKR-SVNSRTCPYFHQ 416
8**************9999988433334699*******************************************8.78888******* PP
trihelix 85 l 85
l
SMil_00025248-RA_Salv 417 L 417
9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00717 | 4.3 | 62 | 119 | IPR001005 | SANT/Myb domain |
| Pfam | PF13837 | 5.1E-8 | 64 | 125 | No hit | No description |
| PROSITE profile | PS50090 | 6.62 | 65 | 117 | IPR017877 | Myb-like domain |
| SMART | SM00717 | 0.025 | 327 | 393 | IPR001005 | SANT/Myb domain |
| CDD | cd12203 | 1.40E-20 | 329 | 398 | No hit | No description |
| Pfam | PF13837 | 6.2E-18 | 329 | 417 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 1.5E-4 | 329 | 392 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50090 | 7.561 | 330 | 391 | IPR017877 | Myb-like domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0001158 | Molecular Function | enhancer sequence-specific DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 437 aa Download sequence Send to blast |
MFDGSEQFHQ FIASSRASLP ISLPFPTLPS FDPYAPPLHH PSSAKKYPLI STNLALESDP 60 STADPWSHDE LLALLKLRST MEIWFPDFTW EHVSRKLGEL GFKRSGEECK EKFEEESCRH 120 FAASKSYRIL SELDEFYPQV PARQGQEDEA ETSKNDEDVV ILRNPEAEAA AEEAEEKGGN 180 HDNNNNNNNS NNRSRSKRKR RDKLETTFKG FCEAVVSKMM AKQEELHRRL MDDVVRREEE 240 KIAREEAWRG REAERARREL EARSKEQKTA AERQATIIDF LKHFSGSSSA SRSPELVDAS 300 NSNVSNSSAS FHPAAPPSPS SGAGAGAGRR WPRDEVQALI KLRCKAGGGG GGGGEEEGGR 360 GPLWERISKG MMEMGYKRSA KRCKEKWENI NKYFRKTRDS SKKRSVNSRT CPYFHQLSCL 420 YSDGTIVAPN RSDAEMN |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 192 | 197 | RSRSKR |
| 2 | 197 | 202 | RKRRDK |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00011 | PBM | Transfer from AT5G28300 | Download |
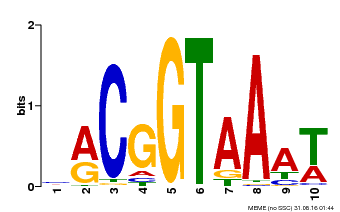
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_022863579.1 | 1e-120 | trihelix transcription factor GTL2 | ||||
| TrEMBL | A0A4D8XPB5 | 1e-143 | A0A4D8XPB5_SALSN; Uncharacterized protein | ||||
| STRING | XP_004495792.1 | 1e-115 | (Cicer arietinum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA13700 | 10 | 13 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G28300.1 | 5e-33 | Trihelix family protein | ||||



