 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | SMil_00026506-RA_Salv | ||||||||
| Common Name | SPL14 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Lamiaceae; Nepetoideae; Mentheae; Salvia
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 228aa MW: 25164.2 Da PI: 8.5977 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 128.5 | 2.5e-40 | 25 | 101 | 2 | 78 |
-SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 2 CqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
C v+gC+adls +++yhrrhkvCe+hsk+p+v++ g+eqrfCqqCsrfh+l efDe+krsCr+rL++hn+rrrk+qa
SMil_00026506-RA_Salv 25 CLVDGCSADLSVCRDYHRRHKVCEAHSKTPKVTIGGREQRFCQQCSRFHSLVEFDEGKRSCRKRLDGHNRRRRKAQA 101
**************************************************************************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 1.8E-33 | 19 | 86 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 31.752 | 22 | 99 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 2.35E-37 | 23 | 102 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 1.3E-30 | 25 | 98 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0048653 | Biological Process | anther development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 228 aa Download sequence Send to blast |
LRSMESSSSS KRAKAPANVA QVAHCLVDGC SADLSVCRDY HRRHKVCEAH SKTPKVTIGG 60 REQRFCQQCS RFHSLVEFDE GKRSCRKRLD GHNRRRRKAQ ARSVTGVEER LVSFSSSAPQ 120 IVAGGGVVGY PWSGVVVKDE NEIGALQQQL NYMDSAASAA HHHHHQHQLS SSPAFVDPNV 180 MMECDGALSL LSSPPPAIID YHQTTSPTLH FPHQNVGSSH QTLSFMWD |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 2e-28 | 25 | 98 | 11 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3' (By similarity). May be involved in panicle development. {ECO:0000250}. | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. {ECO:0000250}. | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00555 | DAP | Transfer from AT5G50570 | Download |
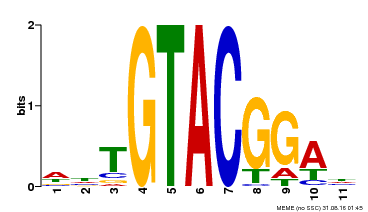
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Negatively regulated by microRNAs miR156 and miR157. {ECO:0000305|PubMed:12202040}. | |||||
| UniProt | INDUCTION: Negatively regulated by microRNAs miR156b and miR156h. {ECO:0000305|PubMed:16861571}. | |||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KF437890 | 0.0 | KF437890.1 Salvia miltiorrhiza SQUAMOSA promoter binding protein-like 14 (SPL14) mRNA, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_001311938.1 | 2e-61 | teosinte glume architecture 1-like | ||||
| Refseq | XP_009621035.1 | 2e-61 | PREDICTED: teosinte glume architecture 1-like | ||||
| Refseq | XP_009621036.1 | 2e-61 | PREDICTED: teosinte glume architecture 1-like | ||||
| Refseq | XP_009621037.1 | 2e-61 | PREDICTED: teosinte glume architecture 1-like | ||||
| Refseq | XP_009621038.1 | 2e-61 | PREDICTED: teosinte glume architecture 1-like | ||||
| Refseq | XP_016443564.1 | 2e-61 | PREDICTED: teosinte glume architecture 1-like isoform X1 | ||||
| Refseq | XP_016443565.1 | 2e-61 | PREDICTED: teosinte glume architecture 1-like isoform X1 | ||||
| Refseq | XP_016443566.1 | 2e-61 | PREDICTED: teosinte glume architecture 1-like isoform X1 | ||||
| Refseq | XP_016443567.1 | 2e-61 | PREDICTED: teosinte glume architecture 1-like isoform X1 | ||||
| Swissprot | B9DI20 | 2e-40 | SP13A_ARATH; Squamosa promoter-binding-like protein 13A | ||||
| Swissprot | P0DI11 | 2e-40 | SP13B_ARATH; Squamosa promoter-binding-like protein 13B | ||||
| Swissprot | Q6YZE8 | 1e-39 | SPL16_ORYSJ; Squamosa promoter-binding-like protein 16 | ||||
| TrEMBL | A0A075FJP6 | 1e-166 | A0A075FJP6_SALMI; SQUAMOSA promoter binding protein-like 14 (Fragment) | ||||
| STRING | XP_009621034.1 | 7e-61 | (Nicotiana tomentosiformis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA4920 | 20 | 37 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G50670.1 | 2e-42 | SBP family protein | ||||



