 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | SMil_00026667-RA_Salv | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Lamiaceae; Nepetoideae; Mentheae; Salvia
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 240aa MW: 27385.2 Da PI: 4.3407 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 60.1 | 3.4e-19 | 52 | 105 | 3 | 56 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
k+++++ +q+++Le+ Fe ++++ e++ +LA++lgL+ rqV vWFqNrRa++k
SMil_00026667-RA_Salv 52 KKRRLSVDQVKALEKNFEVENKLEPERKVKLAQELGLQPRQVAVWFQNRRARWK 105
566899***********************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 130 | 9.2e-42 | 51 | 142 | 1 | 92 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLr 88
ekkrrls +qvk+LE++Fe e+kLeperKv+la+eLglqprqvavWFqnrRAR+ktkqlE+dy Lk++yd lk + erL++++e+L
SMil_00026667-RA_Salv 51 EKKRRLSVDQVKALEKNFEVENKLEPERKVKLAQELGLQPRQVAVWFQNRRARWKTKQLERDYGGLKASYDGLKLNFERLQRDNESLL 138
69*************************************************************************************9 PP
HD-ZIP_I/II 89 eelk 92
+el+
SMil_00026667-RA_Salv 139 KELN 142
9885 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 3.68E-19 | 45 | 109 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.443 | 47 | 107 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 6.3E-18 | 50 | 111 | IPR001356 | Homeobox domain |
| Pfam | PF00046 | 2.2E-16 | 52 | 105 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 5.71E-16 | 52 | 108 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 3.7E-20 | 54 | 113 | IPR009057 | Homeodomain-like |
| PRINTS | PR00031 | 5.7E-6 | 78 | 87 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 82 | 105 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 5.7E-6 | 87 | 103 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 1.4E-14 | 107 | 142 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009788 | Biological Process | negative regulation of abscisic acid-activated signaling pathway | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 240 aa Download sequence Send to blast |
MKRLSNSDSL GGLMSICPNA TEEESNNPMY AREFQTMLEG LEEEESGHVS EKKRRLSVDQ 60 VKALEKNFEV ENKLEPERKV KLAQELGLQP RQVAVWFQNR RARWKTKQLE RDYGGLKASY 120 DGLKLNFERL QRDNESLLKE LNGDDEEKTR LESEIGGAEA EMENPEGGKK EEASWPKFGD 180 GSSDSDSSAI LNEENIGFQL GAPQLVKIEE HNFFGEESCS TLFSDDQPPS LPWYCNDQWD |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 99 | 107 | RRARWKTKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
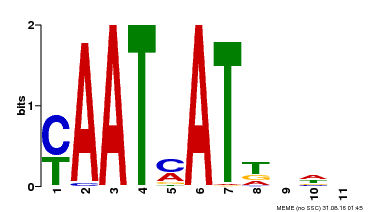
| UniProt | Transcription activator that may act as growth regulators in response to water deficit. Interacts with the core sequence 5'-CAATTATTA-3' of promoters in response to ABA and in an ABI1-dependent manner. Involved in the negative regulation of the ABA signaling pathway. {ECO:0000269|PubMed:10527431, ECO:0000269|PubMed:12065416}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00271 | DAP | Transfer from AT2G22430 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By water deficit, by abscisic acid (ABA) and by salt stress. Self expression regulation. {ECO:0000269|PubMed:10527431, ECO:0000269|PubMed:12065416, ECO:0000269|PubMed:16055682}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011097532.1 | 1e-104 | homeobox-leucine zipper protein ATHB-6 | ||||
| Swissprot | P46668 | 3e-70 | ATHB6_ARATH; Homeobox-leucine zipper protein ATHB-6 | ||||
| TrEMBL | A0A4D9AYZ9 | 1e-104 | A0A4D9AYZ9_SALSN; Homeobox-leucine zipper protein | ||||
| STRING | Migut.L00161.1.p | 1e-101 | (Erythranthe guttata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA2718 | 24 | 53 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G22430.1 | 1e-71 | homeobox protein 6 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



