 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | SapurV1A.0230s0270.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Salicaceae; Saliceae; Salix
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 269aa MW: 30452.6 Da PI: 8.4784 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 51 | 3.3e-16 | 9 | 56 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT+eEd ll ++v+++G g+W I+ + g++R++k+c+ rw++yl
SapurV1A.0230s0270.1.p 9 KGAWTEEEDILLRKCVEKYGEGRWCQIPLKAGLNRCRKSCRMRWLNYL 56
79********************************************97 PP
| |||||||
| 2 | Myb_DNA-binding | 44.7 | 3.1e-14 | 62 | 106 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
rg ++ E +l+++++k+lG++ W++Ia +++ gRt++++k++w++
SapurV1A.0230s0270.1.p 62 RGQFSSGEVDLIIRLHKLLGNR-WSLIAGRLP-GRTANDVKNYWNTN 106
7899999***************.*********.***********986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 24.998 | 4 | 60 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.9E-22 | 6 | 59 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 4.97E-29 | 6 | 103 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 9.4E-16 | 8 | 58 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 3.1E-15 | 9 | 56 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 6.04E-12 | 11 | 56 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 1.5E-22 | 60 | 111 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 7.0E-13 | 61 | 109 | IPR001005 | SANT/Myb domain |
| PROSITE profile | PS51294 | 19.021 | 61 | 111 | IPR017930 | Myb domain |
| Pfam | PF00249 | 7.5E-13 | 62 | 107 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 3.54E-9 | 69 | 107 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0031540 | Biological Process | regulation of anthocyanin biosynthetic process | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 269 aa Download sequence Send to blast |
MTSSFGVRKG AWTEEEDILL RKCVEKYGEG RWCQIPLKAG LNRCRKSCRM RWLNYLKPNI 60 KRGQFSSGEV DLIIRLHKLL GNRWSLIAGR LPGRTANDVK NYWNTNLSKK VVSSTRDAQT 120 KPEAKSITKH NIIKPRPRNF KNLCWLRAGK GTPFINVGSQ YGDDLCKPYS TTAFPPSDIN 180 EIESMWWESL LDDKEINPTN NNSCLGSCSA ANQEPIKNII VEDNPRGGIM IGDVFSEHGQ 240 NCWDDIAFDA DLWNLINTEI DQQGLEGF* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gv2_A | 1e-23 | 9 | 111 | 4 | 105 | MYB PROTO-ONCOGENE PROTEIN |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator, when associated with BHLH002/EGL3/MYC146, BHLH012/MYC1, or BHLH042/TT8. {ECO:0000269|PubMed:15361138}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00213 | DAP | Transfer from AT1G66370 | Download |
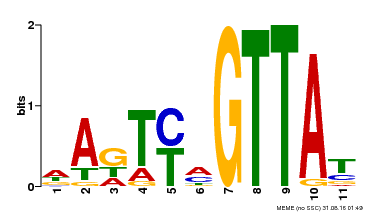
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | SapurV1A.0230s0270.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024444002.1 | 1e-168 | transcription factor MYB90 | ||||
| Swissprot | Q9FNV9 | 4e-64 | MY113_ARATH; Transcription factor MYB113 | ||||
| TrEMBL | U5GFP3 | 1e-166 | U5GFP3_POPTR; Uncharacterized protein | ||||
| STRING | POPTR_0005s14470.1 | 1e-165 | (Populus trichocarpa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF31 | 34 | 817 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G66370.1 | 2e-66 | myb domain protein 113 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | SapurV1A.0230s0270.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



