 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | SapurV1A.0961s0160.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Salicaceae; Saliceae; Salix
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 678aa MW: 76223.5 Da PI: 6.3167 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 139.8 | 2.6e-43 | 76 | 200 | 2 | 134 |
DUF822 2 gsgrkptwkErEnnkrRERrRRaiaakiyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyrkgskpleeaeaagssasa 88
g+g+++++kE+E++k+RER+RRai+++++aGLR++G+++lp+raD+n+Vl+AL+reAGw+ve+DGtt+r++ p ++gs+ +
SapurV1A.0961s0160.1.p 76 GKGKREREKEKERTKLRERHRRAITSRMLAGLRQYGDFPLPARADMNDVLAALAREAGWTVEADGTTFRQSPPPS----HTGSFGVR 158
899******************************************************************776555....88998888 PP
DUF822 89 spesslqsslkssalaspvesysaspksssfpspssldsislasa..a 134
+es+l k++a++ e + ++++ ++++sp slds+ +++ +
SapurV1A.0961s0160.1.p 159 PVESPLL---KNCAVK---ECQPSVLRIDESLSPGSLDSMVISERenS 200
8888866...666655...45667788999999999999999877441 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 1.9E-43 | 76 | 213 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| SuperFamily | SSF51445 | 2.11E-159 | 236 | 671 | IPR017853 | Glycoside hydrolase superfamily |
| Gene3D | G3DSA:3.20.20.80 | 2.2E-172 | 238 | 669 | IPR013781 | Glycoside hydrolase, catalytic domain |
| Pfam | PF01373 | 4.7E-86 | 263 | 635 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 8.0E-56 | 276 | 290 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 8.0E-56 | 297 | 315 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 8.0E-56 | 319 | 340 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 8.0E-56 | 412 | 434 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 8.0E-56 | 485 | 504 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 8.0E-56 | 519 | 535 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 8.0E-56 | 536 | 547 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 8.0E-56 | 554 | 577 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 8.0E-56 | 592 | 614 | IPR001554 | Glycoside hydrolase, family 14 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000272 | Biological Process | polysaccharide catabolic process | ||||
| GO:0048831 | Biological Process | regulation of shoot system development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0016161 | Molecular Function | beta-amylase activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 678 aa Download sequence Send to blast |
MNTNNSNQDL LINPQQTQIS QPDPYSHLPH PVQPGPSPSP HTQTRRPRGF AATAAAAAAA 60 ASADSTPAVS SPSASGKGKR EREKEKERTK LRERHRRAIT SRMLAGLRQY GDFPLPARAD 120 MNDVLAALAR EAGWTVEADG TTFRQSPPPS HTGSFGVRPV ESPLLKNCAV KECQPSVLRI 180 DESLSPGSLD SMVISERENS RNEKYTSASP INSVIECLDA DQLIQDVHSG MHQNDFTENS 240 RVPVYVMLAN GFINNCCHLI DPQGVRQELS HMKSLDVDGV VVECWWGVVE AWSPQKYVWS 300 GYRELFNIIQ EFKLKLQVLM AFHEYGGNDS GDVLISLPQW VLEIGKDNQD IFFTDREGRR 360 NTECLSWGID KERVLKGRTG IEVYFDFMRS FRTEFNDLFT EGLITAIEIG LGPSGELKYP 420 SFSERLGWRY PGIGEFQCYD KYSQQNLRKA AKLRGHSFWA KGPDNAGQYN SRPHETGFFC 480 ERGDYDSYFG RFFLHWYSQS LIDHADNVLS LASFAFEDTK IIVKVPAVYW WYRTASHAAE 540 LTAGYYNPTN QDGYSPVFEV LKKHSVIVKF VCSGLHVSGF ENDEALVDPE GLSWQVLNSA 600 WDRGLTVAGV NMLACYDRDG YRRVVEMAKP RNDPDHHRFS FFVYQQPSAL SHGTICFPEL 660 DYFIKCMHGG ITGDLVS* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wdq_A | 1e-123 | 242 | 631 | 12 | 401 | Beta-amylase |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00543 | DAP | Transfer from AT5G45300 | Download |
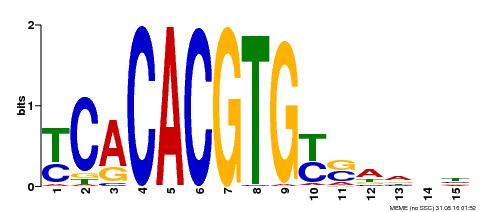
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | SapurV1A.0961s0160.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002304400.2 | 0.0 | beta-amylase 8 isoform X2 | ||||
| Swissprot | Q9FH80 | 0.0 | BAM8_ARATH; Beta-amylase 8 | ||||
| TrEMBL | A0A2K2B4Z1 | 0.0 | A0A2K2B4Z1_POPTR; Beta-amylase | ||||
| STRING | POPTR_0003s10570.1 | 0.0 | (Populus trichocarpa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF11902 | 30 | 32 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G45300.1 | 0.0 | beta-amylase 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | SapurV1A.0961s0160.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



