 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sevir.2G380600.3.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Cenchrinae; Setaria
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 523aa MW: 56695.7 Da PI: 6.5093 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 102.2 | 3e-32 | 140 | 197 | 2 | 60 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS-- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhek 60
+DgynWrKYGqK+vkgse+prsYY+Ct+++C+vkk +ers d++++e++Y+g+Hnh+k
Sevir.2G380600.3.p 140 EDGYNWRKYGQKHVKGSENPRSYYKCTHPNCEVKKLLERSL-DGQITEVVYKGRHNHPK 197
8****************************************.***************85 PP
| |||||||
| 2 | WRKY | 107.6 | 6.1e-34 | 313 | 371 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
ldDgy+WrKYGqK+vkg+++prsYY+Ct+ gCpv+k+ver+++dpk v++tYeg+Hnhe
Sevir.2G380600.3.p 313 LDDGYRWRKYGQKVVKGNPNPRSYYKCTHMGCPVRKHVERASHDPKSVITTYEGKHNHE 371
59********************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 5.3E-28 | 130 | 198 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 23.801 | 134 | 198 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 7.32E-25 | 138 | 198 | IPR003657 | WRKY domain |
| SMART | SM00774 | 3.6E-35 | 139 | 197 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 1.7E-24 | 140 | 196 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 4.6E-38 | 298 | 373 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 4.58E-30 | 305 | 373 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 38.297 | 308 | 373 | IPR003657 | WRKY domain |
| SMART | SM00774 | 5.4E-40 | 313 | 372 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 1.2E-26 | 314 | 371 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009611 | Biological Process | response to wounding | ||||
| GO:0009961 | Biological Process | response to 1-aminocyclopropane-1-carboxylic acid | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 523 aa Download sequence Send to blast |
MAAILNKSTH LDILPSPRDK SAGSGHEDGG SRDFEFKPHL NSQSVAPAVN DPNHRDTSMQ 60 NYTLNHATSS SNLMTESKSL CSRESTHTAL VSSDPKPLAI VGPSDNMPAE VGTSEMNQMN 120 SSENAAQETQ SENVAEKSAE DGYNWRKYGQ KHVKGSENPR SYYKCTHPNC EVKKLLERSL 180 DGQITEVVYK GRHNHPKPQP NRRLANGAVP SSQGEERYDG VAPVEDKPSN IYSNLCNQVH 240 SAGMLDPVPG PASDDDVDAG GGRPYPGDDT NEDDDLDSKR RKMESAGIDA ALMGKPNREP 300 RVVVQTVSEV DILDDGYRWR KYGQKVVKGN PNPRSYYKCT HMGCPVRKHV ERASHDPKSV 360 ITTYEGKHNH EVPASRNASH EMSTAPMKPA VHPINSNMPG LGGMMRACDA RAFTNQYSQA 420 AESDTISLDL GVGISPNHSD ATNQMQPSVP EPMQYQMQHM APVYSSMGLP GMAVATVPGN 480 AASSMYGSRE EKGNEGFTFK AAPLDRSANL CYSSAGNLVM GP* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 2e-36 | 140 | 374 | 8 | 78 | Probable WRKY transcription factor 4 |
| 2lex_A | 2e-36 | 140 | 374 | 8 | 78 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in starch synthesis (PubMed:12953112). Acts as a transcriptional activator in sugar signaling (PubMed:16167901). Interacts specifically with the SURE and W-box elements, but not with the SP8a element (PubMed:12953112). {ECO:0000269|PubMed:12953112, ECO:0000269|PubMed:16167901}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00455 | DAP | Transfer from AT4G26640 | Download |
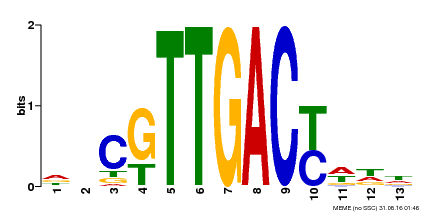
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Sevir.2G380600.3.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated by sugar. {ECO:0000269|PubMed:12953112}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT054888 | 0.0 | BT054888.1 Zea mays full-length cDNA clone ZM_BFc0109P23 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004958143.1 | 0.0 | WRKY transcription factor SUSIBA2 | ||||
| Swissprot | Q6VWJ6 | 0.0 | WRK46_HORVU; WRKY transcription factor SUSIBA2 | ||||
| TrEMBL | K3ZRL5 | 0.0 | K3ZRL5_SETIT; Uncharacterized protein | ||||
| STRING | Si029245m | 0.0 | (Setaria italica) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G26640.1 | 1e-107 | WRKY family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Sevir.2G380600.3.p |



