 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sevir.3G049500.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Cenchrinae; Setaria
|
||||||||
| Family | Trihelix | ||||||||
| Protein Properties | Length: 341aa MW: 38154.3 Da PI: 6.0076 | ||||||||
| Description | Trihelix family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | trihelix | 66.3 | 6.5e-21 | 63 | 130 | 1 | 68 |
trihelix 1 rWtkqevlaLiearremeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkrykkikege 68
+W++ e+ a +++r e+++++ ++k++k+lWe+vs++++ +gf r+p qCk+kw+nl +r+k +++ +
Sevir.3G049500.1.p 63 QWSHAETAAFLAVRAELDHSFLTTKRNKALWEAVSARLQGQGFARTPDQCKSKWKNLVTRFKGTEAAA 130
7*************************************************************998765 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00717 | 0.0047 | 60 | 122 | IPR001005 | SANT/Myb domain |
| CDD | cd12203 | 3.03E-19 | 62 | 127 | No hit | No description |
| Pfam | PF13837 | 5.4E-16 | 63 | 128 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 2.6E-4 | 63 | 121 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50090 | 7.654 | 64 | 120 | IPR017877 | Myb-like domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 341 aa Download sequence Send to blast |
MDPFHHLNTS FSNPYHPLLS PSPPHHPHFP PLPPPPPPPL PPPADPAPQL QPSASLERER 60 LPQWSHAETA AFLAVRAELD HSFLTTKRNK ALWEAVSARL QGQGFARTPD QCKSKWKNLV 120 TRFKGTEAAA AAAAHPPPDA DPASASAAAA QQARQFPFHD EMRRIFDARV ERAQALERKR 180 AKGKDVRTED DEGGGEGEDD DDEEEELEAE VGEDEAGSRV ATEARGGGGA KKRRRKQAAS 240 AARDRSADQG EVEAMLREFM RRQVEMEERW MEAAEAREAE RRAREEEWRN AMVALGEERL 300 ALVRRWRERE DAWRARAEER EERRHQLVAA LLAKLGGDSS * |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 230 | 235 | KKRRRK |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00481 | DAP | Transfer from AT5G01380 | Download |
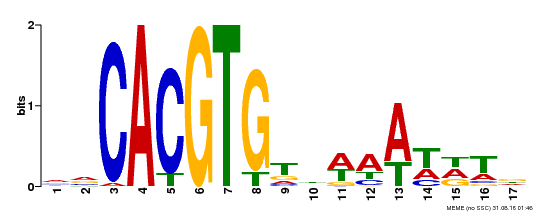
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Sevir.3G049500.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | EU946342 | 4e-95 | EU946342.1 Zea mays clone 305005 mRNA sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_022680762.1 | 1e-117 | trihelix transcription factor GT-3b-like | ||||
| TrEMBL | A0A368QDJ5 | 1e-115 | A0A368QDJ5_SETIT; Uncharacterized protein | ||||
| STRING | ORUFI05G02060.1 | 1e-104 | (Oryza rufipogon) | ||||
| STRING | OS05T0128000-00 | 1e-104 | (Oryza sativa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP12595 | 31 | 34 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G01380.1 | 1e-28 | Trihelix family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Sevir.3G049500.1.p |



