 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sevir.4G059600.2.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Cenchrinae; Setaria
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 245aa MW: 26297.2 Da PI: 7.3492 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 51.6 | 1.7e-16 | 115 | 161 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn E+rRR++iN+++ L++l+P++ K +Ka++L +A+eY+k+Lq
Sevir.4G059600.2.p 115 VHNLSEKRRRSKINEKMKALQSLIPNS-----NKTDKASMLDEAIEYLKQLQ 161
6*************************7.....5******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.280.10 | 2.4E-20 | 108 | 171 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 7.2E-21 | 108 | 172 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 18.076 | 111 | 160 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 6.51E-18 | 114 | 165 | No hit | No description |
| Pfam | PF00010 | 6.0E-14 | 115 | 161 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 2.9E-18 | 117 | 166 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0007623 | Biological Process | circadian rhythm | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0010114 | Biological Process | response to red light | ||||
| GO:0010154 | Biological Process | fruit development | ||||
| GO:0010187 | Biological Process | negative regulation of seed germination | ||||
| GO:0048440 | Biological Process | carpel development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 245 aa Download sequence Send to blast |
MDEQGLGGFG GGGRGAVRDH GREAMALLQH QQHQQRRRQL EEEEEEAAAR RQMFAGVAAF 60 PAAAALGLGH GGQQVDYGEE ADGLGDSDAG GSEPEPAQAR QRGGSGSKRS RAAEVHNLSE 120 KRRRSKINEK MKALQSLIPN SNKTDKASML DEAIEYLKQL QLQVQMLSMR NGVYLNPPYL 180 SGALEPAQAS QMFAALGGGH ITASSSGAVM PPVNQSSGAH QNINFNWNRH SPTFEHFGCL 240 NLLR* |
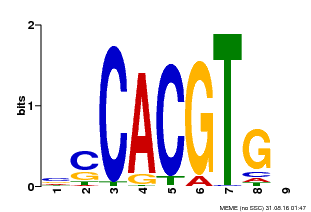
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00035 | PBM | Transfer from AT4G36930 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Sevir.4G059600.2.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT035360 | 1e-177 | BT035360.1 Zea mays full-length cDNA clone ZM_BFb0057H06 mRNA, complete cds. | |||
| GenBank | KJ726860 | 1e-177 | KJ726860.1 Zea mays clone pUT3401 bHLH transcription factor (bHLH125) mRNA, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025811540.1 | 1e-100 | uncharacterized protein LOC112889235 isoform X1 | ||||
| TrEMBL | A0A2T7E2L4 | 1e-101 | A0A2T7E2L4_9POAL; Uncharacterized protein | ||||
| STRING | GRMZM2G017349_P01 | 5e-99 | (Zea mays) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G36930.1 | 4e-36 | bHLH family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Sevir.4G059600.2.p |



