 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sevir.5G077800.3.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Cenchrinae; Setaria
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 761aa MW: 82331 Da PI: 5.2207 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 69.4 | 4.4e-22 | 54 | 109 | 1 | 56 |
TT--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 1 rrkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+++ +++t++q++eLe+lF+++++p++++r eL+++lgL+ rqVk+WFqNrR+++k
Sevir.5G077800.3.p 54 KKRYHRHTPQQIQELEALFKECPHPDEKQRGELSRRLGLDPRQVKFWFQNRRTQMK 109
678899***********************************************999 PP
| |||||||
| 2 | START | 190 | 1.2e-59 | 270 | 502 | 1 | 206 |
HHHHHHHHHHHHHHHHC-TT-EEEE......EXCCTTEEEEEEESSS......SCEEEEEEEECCS.CHHHHHHHHHCCCGGCT-TT-S.. CS
START 1 elaeeaaqelvkkalaeepgWvkss......esengdevlqkfeeskv.....dsgealrasgvvd.mvlallveellddkeqWdetla.. 77
ela +a++elvk+a+ +ep+W s e++n +e+ q+f + + + +ea+r+sg+v+ ++++ lve+l+d + +W+ ++
Sevir.5G077800.3.p 270 ELAISAMDELVKLAQIDEPLWLPSLngspnkEMLNFEEYAQSFLPCIGvkpvgYVSEASRESGLVIiDDSVALVETLMDER-RWSDMFScm 359
57889********************999999999*********998889**************997245679*********.********* PP
..EEEEEEEECTT......EEEEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE..-TTS--....-TTSEE-EESSEEEEE CS
START 78 ..kaetlevissg......galqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvd..seqkppe...sssvvRaellpSgili 154
ka++le ++sg g l lm+aelq+lsplvp R+++f+R+++ql +g w++vdvS+d ++++++ +++ vR+++lpSg+++
Sevir.5G077800.3.p 360 iaKATILEEVTSGiagsrnGMLLLMKAELQVLSPLVPiREVTFLRFCKQLAEGAWAVVDVSIDglVRDQNSAtisNAGNVRCRRLPSGCVM 450
*9************************************************************95433444446899*************** PP
EEECTCEEEEEEEE-EE--SSXXHHHHHHHHHHHHHHHHHHHHHHTXXXXXX CS
START 155 epksnghskvtwvehvdlkgrlphwllrslvksglaegaktwvatlqrqcek 206
++++ng++kvtwveh++++++++h+l+r+l++sgla+ga++w+a lqrqce+
Sevir.5G077800.3.p 451 QDTPNGYCKVTWVEHTEYDEASVHQLYRPLLRSGLAFGARRWLAMLQRQCEC 502
**************************************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 6.68E-22 | 35 | 111 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 6.9E-24 | 39 | 111 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 18.285 | 51 | 111 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 4.8E-21 | 52 | 115 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 6.10E-20 | 53 | 111 | No hit | No description |
| Pfam | PF00046 | 1.4E-19 | 54 | 109 | IPR001356 | Homeobox domain |
| PROSITE pattern | PS00027 | 0 | 86 | 109 | IPR017970 | Homeobox, conserved site |
| PROSITE profile | PS50848 | 47.171 | 261 | 505 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 1.37E-31 | 263 | 502 | No hit | No description |
| CDD | cd08875 | 5.82E-115 | 265 | 501 | No hit | No description |
| SMART | SM00234 | 1.7E-48 | 270 | 502 | IPR002913 | START domain |
| Pfam | PF01852 | 4.6E-52 | 271 | 502 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 8.24E-22 | 522 | 730 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009827 | Biological Process | plant-type cell wall modification | ||||
| GO:0042335 | Biological Process | cuticle development | ||||
| GO:0043481 | Biological Process | anthocyanin accumulation in tissues in response to UV light | ||||
| GO:0048765 | Biological Process | root hair cell differentiation | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 761 aa Download sequence Send to blast |
MGGGRALPGG AGDGSARDAD AENDSRSGSD HLDAMSGGGE DEDDAEPGNP RKRKKRYHRH 60 TPQQIQELEA LFKECPHPDE KQRGELSRRL GLDPRQVKFW FQNRRTQMKT QLERHENALL 120 KQENDKLRAE NMTIREAMRT PMCGSCGSPA MLGEVSLEEQ HLCIENARLK DELNRVYALA 180 TKFLGKPMAM LSGPMLQPHL SSLPMPSSSL ELAVGGFRGL GSIPSATMPG SMSEFAGGVS 240 SPLGTVITPA RATGSAPPSM VGIDRSMLLE LAISAMDELV KLAQIDEPLW LPSLNGSPNK 300 EMLNFEEYAQ SFLPCIGVKP VGYVSEASRE SGLVIIDDSV ALVETLMDER RWSDMFSCMI 360 AKATILEEVT SGIAGSRNGM LLLMKAELQV LSPLVPIREV TFLRFCKQLA EGAWAVVDVS 420 IDGLVRDQNS ATISNAGNVR CRRLPSGCVM QDTPNGYCKV TWVEHTEYDE ASVHQLYRPL 480 LRSGLAFGAR RWLAMLQRQC ECLAILMSPD TVSANDSSVI TQEGKRSMLK LARRMTENFC 540 AGVSASSARE WSKLDGATGS IGEDVRVMAR KSVDEPGEPP GVVLSAATSV WVPVAPEKLF 600 NFLRDEQLRA EWDILSNGGP MQEMANIAKG QEHGNSVSLL RASAMSANQS SMLILQETCT 660 DASGSMVVYA PVDIPAMQLV MNGGDSTYVA LLPSGFAILP DGPSTGADHK TGGSLLTVAF 720 QILVNSQPTA KLTVESVETV NNLISCTIKK IKTALQWDSA * |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 49 | 55 | PRKRKKR |
| 2 | 50 | 54 | RKRKK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00421 | DAP | Transfer from AT4G00730 | Download |
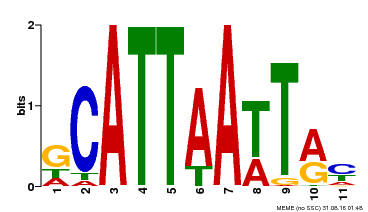
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Sevir.5G077800.3.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT063249 | 0.0 | BT063249.1 Zea mays full-length cDNA clone ZM_BFc0042P14 mRNA, complete cds. | |||
| GenBank | BT064453 | 0.0 | BT064453.1 Zea mays full-length cDNA clone ZM_BFc0170E20 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004968001.1 | 0.0 | homeobox-leucine zipper protein ROC5 isoform X1 | ||||
| Refseq | XP_004968002.1 | 0.0 | homeobox-leucine zipper protein ROC5 isoform X2 | ||||
| Swissprot | Q6EPF0 | 0.0 | ROC5_ORYSJ; Homeobox-leucine zipper protein ROC5 | ||||
| TrEMBL | K3XEM7 | 0.0 | K3XEM7_SETIT; Uncharacterized protein | ||||
| TrEMBL | K3XEN0 | 0.0 | K3XEN0_SETIT; Uncharacterized protein | ||||
| STRING | Si000344m | 0.0 | (Setaria italica) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G61150.1 | 0.0 | homeodomain GLABROUS 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Sevir.5G077800.3.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



