 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sevir.9G316400.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Cenchrinae; Setaria
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 707aa MW: 75291.2 Da PI: 6.6087 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 39 | 1.4e-12 | 528 | 573 | 4 | 54 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksL 54
+h e+Er+RR+++N++f Lr ++P+ K++Ka+ L A+ YI++L
Sevir.9G316400.1.p 528 NHVEAERQRREKLNQRFYALRAVVPNV-----SKMDKASLLGDAISYINEL 573
799***********************6.....5***************998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF14215 | 1.6E-53 | 59 | 239 | IPR025610 | Transcription factor MYC/MYB N-terminal |
| PROSITE profile | PS50888 | 17.39 | 524 | 573 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 3.32E-14 | 527 | 577 | No hit | No description |
| SuperFamily | SSF47459 | 1.83E-18 | 527 | 595 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 1.4E-18 | 528 | 595 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 4.7E-10 | 528 | 573 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 3.6E-16 | 530 | 579 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009269 | Biological Process | response to desiccation | ||||
| GO:0009611 | Biological Process | response to wounding | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009867 | Biological Process | jasmonic acid mediated signaling pathway | ||||
| GO:0009963 | Biological Process | positive regulation of flavonoid biosynthetic process | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0043619 | Biological Process | regulation of transcription from RNA polymerase II promoter in response to oxidative stress | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0051090 | Biological Process | regulation of sequence-specific DNA binding transcription factor activity | ||||
| GO:2000068 | Biological Process | regulation of defense response to insect | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 707 aa Download sequence Send to blast |
MNLWTDDNAS MMEAFMASAD LPAFPWGAPA GGGASSAAAT PPPPQMPAAM APGFNQDTLQ 60 QRLQAMIEGS RETWTYAIFW QSSVDAATGA SLLGWGDGYY KGCDEDKRKQ KPLTPAAQAE 120 QEHRKRVLRE LNSLISGAAA APDEAVEEEV TDTEWFFLVS MTQSFLNGSG LPGQALFAGQ 180 PTWIASGLSS APCERARQAY NFGLRTMVCV PVGTGVLELG STDVVFQTAE SMAKIRSLFG 240 GGGGTGGGSW PPVQPPAPPP QQPAAGADQA ETDPSVLWLA DAPVMDIKES LSHPSAEISV 300 SKPPPPPQIH FENGSSSTLT ENPSPSVHAP PPPPAPAAAP PQRQHQHNQA HQGPFRRELN 360 FSEFASNPSM AAAPPFFKPE SGEILSFGAD SNGRRNPSPA PPAATASLTT APGSLFSQHT 420 ATLTAAPAND TKNNNNKRSM EATSRASNTN YHPAATANEG MLSFSSAPTT RPSTGTGAPA 480 KSESDHSDLD ASVREVESSR VVAPPPEAEK RPRKRGRKPA NGREEPLNHV EAERQRREKL 540 NQRFYALRAV VPNVSKMDKA SLLGDAISYI NELRGKLTSL ESDKDTLHAQ IEALKKERDA 600 RPAPHAAGLG GHDAGPRCHA VEIDAKILGL EAMIRVQCHK RNHPSARLMT ALRELDLDVY 660 HASVSVVKDL MIQQVAVKMA SRVYSQEQLN AALYSRLAEP GTAMGR* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4rqw_A | 2e-63 | 52 | 239 | 2 | 192 | Transcription factor MYC3 |
| 4rqw_B | 2e-63 | 52 | 239 | 2 | 192 | Transcription factor MYC3 |
| 4rs9_A | 2e-63 | 52 | 239 | 2 | 192 | Transcription factor MYC3 |
| 4yz6_A | 2e-63 | 52 | 239 | 2 | 192 | Transcription factor MYC3 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 509 | 517 | KRPRKRGRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator involved in jasmonate (JA) signaling pathway during spikelet development. Binds to the G2 region G-box (5'-CACGTG-3') of the MADS1 promoter and thus directly regulates the expression of MADS1. Its function in MADS1 activation is abolished by TIFY3/JAZ1 which directly target MYC2 during spikelet development. {ECO:0000269|PubMed:24647160}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00084 | PBM | Transfer from AT1G32640 | Download |
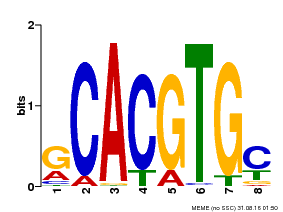
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Sevir.9G316400.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AF061107 | 0.0 | AF061107.1 Zea mays transcription factor MYC7E mRNA, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_012698381.1 | 0.0 | LOW QUALITY PROTEIN: transcription factor MYC2-like | ||||
| Swissprot | Q336P5 | 0.0 | MYC2_ORYSJ; Transcription factor MYC2 | ||||
| TrEMBL | K4AM38 | 0.0 | K4AM38_SETIT; Uncharacterized protein | ||||
| STRING | Si039973m | 0.0 | (Setaria italica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP4452 | 31 | 57 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G32640.1 | 1e-143 | bHLH family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Sevir.9G316400.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



