 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sme2.5_00142.1_g00013.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Solaneae; Solanum
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 884aa MW: 98869.1 Da PI: 4.5439 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 164.2 | 4.8e-51 | 25 | 152 | 1 | 128 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLp..kkvkaeekewyfFskrdkkyatgkrknratksgyWk 84
l+pGfrFhPtdeelv +yL++k+ gk +++ e+++e+d+yk+ePw+L+ +++k+++ ewyfFs+ d+ky +g+r nrat +gyWk
Sme2.5_00142.1_g00013.1 25 LAPGFRFHPTDEELVRYYLRRKACGKLFRF-EAVAEIDVYKSEPWELKvySALKTRDLEWYFFSPVDRKYGNGSRLNRATGQGYWK 109
579************************999.89**************95336777888**************************** PP
NAM 85 atgkdkevlskkgelvglkktLvfykgrapkgektdWvmheyrl 128
atgkd++v + + +++g+kktLvf++grap+g++t+Wvmheyrl
Sme2.5_00142.1_g00013.1 110 ATGKDRPVRH-NAKTIGMKKTLVFHNGRAPDGQRTNWVMHEYRL 152
**********.999****************************98 PP
| |||||||
| 2 | NAM | 106.1 | 4.5e-33 | 375 | 478 | 33 | 128 |
NAM 33 v.ikevdiykvePwdLpk.......kvkaeekewyfFskrdkkyatgkrknratksgyWkatgkdkevlskkgelvglkktLvfyk 110
+ +++d+++ e +dL++ ++k+++ ewyfFs dkky +g r+nrat++gyWk+tgkd++v+++k+e+vg+kktLv++
Sme2.5_00142.1_g00013.1 375 IlQSRIDLLNKENEDLRRlvrggmsRLKTRDLEWYFFSVLDKKYGNGARTNRATEKGYWKTTGKDRAVHRNKSEVVGMKKTLVYHI 460
31456699999999998557787777777888****************************************************** PP
NAM 111 grapkgektdWvmheyrl 128
grapkg++t+Wvmheyrl
Sme2.5_00142.1_g00013.1 461 GRAPKGQRTNWVMHEYRL 478
****************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 2.35E-59 | 19 | 175 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 57.424 | 25 | 175 | IPR003441 | NAC domain |
| Pfam | PF02365 | 1.7E-27 | 27 | 152 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 36.791 | 337 | 501 | IPR003441 | NAC domain |
| Pfam | PF02365 | 2.2E-15 | 396 | 478 | IPR003441 | NAC domain |
| SuperFamily | SSF101941 | 4.05E-38 | 399 | 501 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009555 | Biological Process | pollen development | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 884 aa Download sequence Send to blast |
MEQEGVLVLK PAPARAVAAA PPTSLAPGFR FHPTDEELVR YYLRRKACGK LFRFEAVAEI 60 DVYKSEPWEL KVYSALKTRD LEWYFFSPVD RKYGNGSRLN RATGQGYWKA TGKDRPVRHN 120 AKTIGMKKTL VFHNGRAPDG QRTNWVMHEY RLADEELELA GVVQDAFVLC RIFQKSGLGP 180 PNGDRYAPFI EEEWDDDDTT HLVPGGAAEV DVAIGDEARI EGNDLDQDAR GKAPCQSENL 240 LEARALSFVC KRERSEEELE PLSLAQSKRS KHDDPSSSHA NGSEDSTTSQ QDPPTTMMTT 300 NYSPPLLTFP LLESLEPKEA QPSNSLNFDS SNLEKSVPPG YLKFISNLEN EILNVSMERE 360 TAKIEVMRAQ AMINILQSRI DLLNKENEDL RRLVRGGMSR LKTRDLEWYF FSVLDKKYGN 420 GARTNRATEK GYWKTTGKDR AVHRNKSEVV GMKKTLVYHI GRAPKGQRTN WVMHEYRLID 480 EALDRAGIPQ DAFVLCRVFQ KSGAGPKNGE QYGAPFIEEE WEDDDVQVLP KDEAAEEFVF 540 GDDIHLDANE LDQILGTGTA VAGAAHPSDY SWDENGSTEE TNNSSDAQNW VKQPGELKPI 600 DLAVQQKFDA PDQVNHEYIG ESSKNMNSED GDYLLNEPIV NGTDDLQFSD EAFLEANDLS 660 NPVEVDRSGF DMLEEYLTYF DANDDFQNMS FDPSVFFGND DQISDQALPP EKDIGEATQQ 720 FIAPSEGLGE DTNDLAASSK LVPTKPGSDC QYLFMKKASQ MLGNIHAPPA FASEFPTKEA 780 TLRLNLPSQS VSSVQFTADL VQIRDMSAAG NEMGWPLGKH SDYNIILSFG LSRGGSDDSG 840 MESFHSIHPG KTASAIFRGW FYYLFFSFLM LLMSIRIGTM ISAS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 3swm_A | 1e-47 | 24 | 507 | 19 | 174 | NAC domain-containing protein 19 |
| 3swm_B | 1e-47 | 24 | 507 | 19 | 174 | NAC domain-containing protein 19 |
| 3swm_C | 1e-47 | 24 | 507 | 19 | 174 | NAC domain-containing protein 19 |
| 3swm_D | 1e-47 | 24 | 507 | 19 | 174 | NAC domain-containing protein 19 |
| 3swp_A | 1e-47 | 24 | 507 | 19 | 174 | NAC domain-containing protein 19 |
| 3swp_B | 1e-47 | 24 | 507 | 19 | 174 | NAC domain-containing protein 19 |
| 3swp_C | 1e-47 | 24 | 507 | 19 | 174 | NAC domain-containing protein 19 |
| 3swp_D | 1e-47 | 24 | 507 | 19 | 174 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional repressor that binds to the motif 5'-(C/T)A(C/A)G-3' in the promoter of target genes (PubMed:25578968). Binds also to the 5'-CTTGNNNNNCAAG-3' consensus sequence in chromatin (PubMed:26617990). Can bind to the mitochondrial dysfunction motif (MDM) present in the upstream regions of mitochondrial dysfunction stimulon (MDS) genes involved in mitochondrial retrograde regulation (MRR) (PubMed:24045019). Together with NAC050 and JMJ14, regulates gene expression and flowering time by associating with the histone demethylase JMJ14, probably by the promotion of RNA-mediated gene silencing (PubMed:25578968, PubMed:26617990). Regulates siRNA-dependent post-transcriptional gene silencing (PTGS) through SGS3 expression modulation (PubMed:28207953). Required during pollen development (PubMed:19237690). {ECO:0000269|PubMed:19237690, ECO:0000269|PubMed:24045019, ECO:0000269|PubMed:25578968, ECO:0000269|PubMed:26617990, ECO:0000269|PubMed:28207953}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00612 | ChIP-seq | Transfer from AT3G10490 | Download |
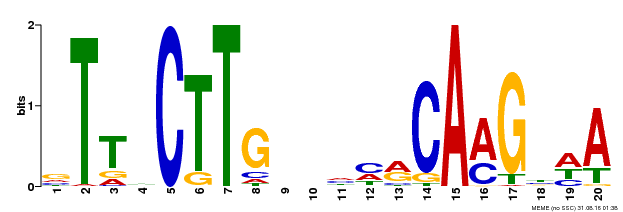
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by type III effector proteins (TTEs) secreted by the pathogenic bacteria P.syringae pv. tomato DC3000 during basal defense. {ECO:0000269|PubMed:16553893}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT013747 | 0.0 | BT013747.1 Lycopersicon esculentum clone 132610F, mRNA sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006361710.1 | 0.0 | PREDICTED: NAC domain-containing protein 53-like | ||||
| Swissprot | Q9SQY0 | 1e-101 | NAC52_ARATH; NAC domain containing protein 52 | ||||
| TrEMBL | A0A3Q7JIC2 | 0.0 | A0A3Q7JIC2_SOLLC; Uncharacterized protein | ||||
| STRING | PGSC0003DMT400079789 | 0.0 | (Solanum tuberosum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA3140 | 24 | 48 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G10500.1 | 3e-96 | NAC domain containing protein 53 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



