 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sme2.5_01440.1_g00007.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Solaneae; Solanum
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 955aa MW: 104964 Da PI: 6.9948 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 53 | 5.8e-17 | 53 | 111 | 3 | 57 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHC....TS-HHHHHHHHHHHHHHHHC CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkkl....gLterqVkvWFqNrRakekk 57
k ++t+eq+e+Le+l++++++ps +r++ ++ +++ rq+kvWFqNrR +ek+
Sme2.5_01440.1_g00007.1 53 KYVRYTPEQVEALERLYHECPKPSSMRRQQFIREYpilsHIEPRQIKVWFQNRRCREKQ 111
5679******************************99*********************97 PP
| |||||||
| 2 | START | 134.9 | 9e-43 | 263 | 437 | 29 | 204 |
CTTEEEEEEESSS.SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-SEEEEEEEECTT..EEEEEEEEXXTTXX-SSX.EEEEE CS
START 29 ngdevlqkfeeskvdsgealrasgvvdmvlallveellddkeqWdetlakaetlevissg..galqlmvaelqalsplvp.Rdfvf 111
+g++++ +++ s++++g a+ra+g+v +++ d++ W ++++++e+l+ + ++ g+++l +++l+a+++l+p Rdf+
Sme2.5_01440.1_g00007.1 263 PGPDSIGIIAISHGCTGMAARACGLVGLDPT------RDRP-SWYRDCRAVEVLNMLPTAngGTIELLYMQLYAPTTLAPsRDFWL 341
57899999***********************......8999.****************9999************************ PP
EEEEEE.TTS-EEEEEEEEE-TTS--....-TTSEE-EESSEEEEEEEECTCEEEEEEEE-EE--SSXXHHHHHHHHHHHHHHHHH CS
START 112 vRyirqlgagdwvivdvSvdseqkppe...sssvvRaellpSgiliepksnghskvtwvehvdlkgrlphwllrslvksglaegak 194
+Ry+ +g++v++++S+ ++q+ p+ +++vRae+lpSg+li+p+++g+s v++v+h++l++++++++lr+l++s+++ ++k
Sme2.5_01440.1_g00007.1 342 LRYTTVTDDGSFVVCERSLGNTQNGPSmphVQNFVRAEMLPSGYLIRPCEGGGSIVHIVDHMNLGAWSVPEVLRPLYESSAVLAQK 427
**************************9988899***************************************************** PP
HHHHHTXXXX CS
START 195 twvatlqrqc 204
t++a+l++++
Sme2.5_01440.1_g00007.1 428 TTMAALRQLR 437
******9986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50071 | 15.418 | 48 | 112 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 8.7E-14 | 50 | 116 | IPR001356 | Homeobox domain |
| SuperFamily | SSF46689 | 4.28E-15 | 50 | 114 | IPR009057 | Homeodomain-like |
| CDD | cd00086 | 9.94E-15 | 53 | 113 | No hit | No description |
| Pfam | PF00046 | 1.4E-14 | 54 | 111 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 3.0E-17 | 55 | 111 | IPR009057 | Homeodomain-like |
| CDD | cd14686 | 1.29E-5 | 105 | 144 | No hit | No description |
| PROSITE profile | PS50848 | 22.227 | 187 | 435 | IPR002913 | START domain |
| SMART | SM00234 | 4.9E-25 | 196 | 439 | IPR002913 | START domain |
| Pfam | PF01852 | 5.1E-40 | 263 | 437 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 4.67E-28 | 263 | 437 | No hit | No description |
| Gene3D | G3DSA:3.30.530.20 | 6.0E-20 | 292 | 435 | IPR023393 | START-like domain |
| Pfam | PF08670 | 1.3E-43 | 751 | 881 | IPR013978 | MEKHLA |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009965 | Biological Process | leaf morphogenesis | ||||
| GO:0010014 | Biological Process | meristem initiation | ||||
| GO:0010075 | Biological Process | regulation of meristem growth | ||||
| GO:0010087 | Biological Process | phloem or xylem histogenesis | ||||
| GO:0048263 | Biological Process | determination of dorsal identity | ||||
| GO:0080060 | Biological Process | integument development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 955 aa Download sequence Send to blast |
MLAESSSGEE LIDKSSVLPH TDHLVRGSLG KSAMVLQVPA TPRDGKSLED NGKYVRYTPE 60 QVEALERLYH ECPKPSSMRR QQFIREYPIL SHIEPRQIKV WFQNRRCREK QRKESSRLQG 120 VNRKLSAMNK LLMEENDRLQ KQVSQLVYEN GYFRKQTQTT KLASKDTSCE SVVTSSQHQL 180 TPQHPPRDAS PAGLLSIAEE TLTEFLSKAT GTAVEWVQMP GMKGCRGGRV KELLCVDYGL 240 ISLRLFSILP HECLGLAEPV GVPGPDSIGI IAISHGCTGM AARACGLVGL DPTRDRPSWY 300 RDCRAVEVLN MLPTANGGTI ELLYMQLYAP TTLAPSRDFW LLRYTTVTDD GSFVVCERSL 360 GNTQNGPSMP HVQNFVRAEM LPSGYLIRPC EGGGSIVHIV DHMNLGAWSV PEVLRPLYES 420 SAVLAQKTTM AALRQLRQLT LEVSQPNVTN WGRRPAALRA LSKRLSRGFN EALSGFSSEG 480 WSTLDNDGMD DVTILVNSSP DKLMGLNLSF SDGFTSLSNA VLCARASMLL QSVTPAILLR 540 FLREHRSEWV DNNIDAYSAA AVKVGPCSLP GVRVSNFGGQ LLEVIKLEGV CHSPEDVIMP 600 RDMFLLQLCS GMDENAVGTC AELIFAPIDA SFADDAPLLP SGFRIIPLDS AKEASSPNRT 660 LDLTSALETG PVGSKVANDL KSTAGTSKSV MTIAFQFAFE SHMQENVASM ARKYVRSFIS 720 SVQRVALALS PSHYGSLGGL RLPLGTPEAH TLAQWIGQSY RHFLGVELLK LGSEGSESIL 780 DSLWHHSDAI ICCSAKALPV FTFANQGGLD MLETTLVALQ DISLEKIFDE HGRKNLCSEF 840 PQIMQQGFAC LQGGICLSSM GRPVSYEKAV AWKVLNEEDV AHYMVYEENV ASMARKYVRS 900 FISSVQRVAL ALSPSHYGSL GGLRLPLGTP EAHTLAQWIL IKNDLRDRKN DFTPI |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in the regulation of meristem development to promote lateral organ formation. May regulates procambial and vascular tissue formation or maintenance, and vascular development in inflorescence stems. {ECO:0000269|PubMed:15598805, ECO:0000269|PubMed:15705957, ECO:0000269|PubMed:16617092}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00200 | DAP | Transfer from AT1G52150 | Download |
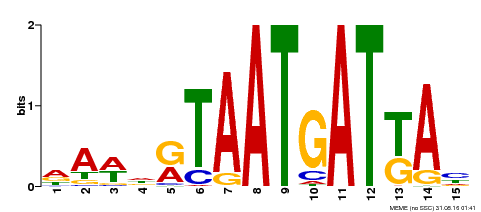
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By auxin. Repressed by miR165 and miR166. {ECO:0000269|PubMed:15773855, ECO:0000269|PubMed:16033795, ECO:0000269|PubMed:16617092, ECO:0000269|PubMed:17237362}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004252354.1 | 0.0 | homeobox-leucine zipper protein ATHB-15 | ||||
| Swissprot | Q9ZU11 | 0.0 | ATB15_ARATH; Homeobox-leucine zipper protein ATHB-15 | ||||
| TrEMBL | A0A3Q7JAS2 | 0.0 | A0A3Q7JAS2_SOLLC; Uncharacterized protein | ||||
| STRING | Solyc12g044410.1.1 | 0.0 | (Solanum lycopersicum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA457 | 24 | 140 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G52150.1 | 0.0 | HD-ZIP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



