 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sme2.5_01517.1_g00008.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Solaneae; Solanum
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 348aa MW: 39782.9 Da PI: 4.7278 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 57.2 | 2.9e-18 | 84 | 138 | 3 | 57 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHHC CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakekk 57
k++++ +q+++Le+ Fe ++++ e++ +LA++lgL+ rqV +WFqNrRa++k+
Sme2.5_01517.1_g00008.1 84 KKRRLRVDQVKALEKNFEVDNKLEPERKVKLARELGLQPRQVAIWFQNRRARWKN 138
45678899*********************************************95 PP
| |||||||
| 2 | HD-ZIP_I/II | 123.9 | 7.7e-40 | 83 | 175 | 1 | 93 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekevee 86
ekkrrl +qvk+LE++Fe ++kLeperKv+lareLglqprqva+WFqnrRAR+k+kqlE dy+ Lk+++d+lk++ e+L++++e+
Sme2.5_01517.1_g00008.1 83 EKKRRLRVDQVKALEKNFEVDNKLEPERKVKLARELGLQPRQVAIWFQNRRARWKNKQLEIDYNLLKANFDSLKHNYEALKHDNEA 168
69***********************************************************************************9 PP
HD-ZIP_I/II 87 Lreelke 93
+ +e++e
Sme2.5_01517.1_g00008.1 169 ILKEINE 175
9999876 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50071 | 17.151 | 79 | 139 | IPR001356 | Homeobox domain |
| SuperFamily | SSF46689 | 7.7E-19 | 81 | 141 | IPR009057 | Homeodomain-like |
| SMART | SM00389 | 2.0E-17 | 82 | 143 | IPR001356 | Homeobox domain |
| Pfam | PF00046 | 1.5E-15 | 84 | 138 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 7.06E-16 | 84 | 140 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 3.6E-19 | 91 | 139 | IPR009057 | Homeodomain-like |
| PRINTS | PR00031 | 1.0E-5 | 110 | 119 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 114 | 137 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 1.0E-5 | 119 | 135 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 1.7E-14 | 139 | 180 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009637 | Biological Process | response to blue light | ||||
| GO:0030308 | Biological Process | negative regulation of cell growth | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0048510 | Biological Process | regulation of timing of transition from vegetative to reproductive phase | ||||
| GO:0048573 | Biological Process | photoperiodism, flowering | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 348 aa Download sequence Send to blast |
MIWTCAEKKY RCSCYEVREV GFRSYTEGQR FCDESFQKQP LYLGSDEHKN WNKHVYSRDF 60 QSMLDGLDDE EATCVEESGV GPEKKRRLRV DQVKALEKNF EVDNKLEPER KVKLARELGL 120 QPRQVAIWFQ NRRARWKNKQ LEIDYNLLKA NFDSLKHNYE ALKHDNEAIL KEINELKSKL 180 YGGNNESKVS VKEEALESET DDKEAEQSKQ YNNSNSTELN FDRIIMNPSI IFTDFKDGSS 240 DSDSSAILNE DNAAISSSGA FLIKSGSRSG SGSGSGSSSP NSNSSLNCFQ FLESNPKSIL 300 ADEVINFSSS QFVKMEEHNF FAGEECCSTL FSDEQAPTLH WYCPDDWN |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 131 | 139 | RRARWKNKQ |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00051 | PBM | Transfer from AT4G40060 | Download |
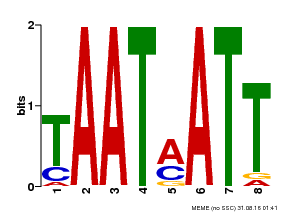
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006366219.1 | 1e-166 | PREDICTED: homeobox-leucine zipper protein ATHB-6-like | ||||
| TrEMBL | M1BEC0 | 1e-164 | M1BEC0_SOLTU; Uncharacterized protein | ||||
| STRING | PGSC0003DMT400043251 | 1e-165 | (Solanum tuberosum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA2718 | 24 | 53 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G40060.1 | 7e-60 | homeobox protein 16 | ||||



