 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sme2.5_02242.1_g00002.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Solaneae; Solanum
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 728aa MW: 77883.7 Da PI: 7.4535 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 49.9 | 5.6e-16 | 451 | 497 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn ErrRRdriN+++ L+el+P++ K +Ka++L +A+eY+k Lq
Sme2.5_02242.1_g00002.1 451 VHNLSERRRRDRINEKMRALQELIPNC-----NKADKASMLDEAIEYLKTLQ 497
6*************************8.....6******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00083 | 4.60E-17 | 443 | 501 | No hit | No description |
| SuperFamily | SSF47459 | 1.96E-20 | 444 | 508 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 2.9E-20 | 444 | 505 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 18.228 | 447 | 496 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 2.5E-13 | 451 | 497 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 2.1E-17 | 453 | 502 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0009740 | Biological Process | gibberellic acid mediated signaling pathway | ||||
| GO:0010017 | Biological Process | red or far-red light signaling pathway | ||||
| GO:0031539 | Biological Process | positive regulation of anthocyanin metabolic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 728 aa Download sequence Send to blast |
MARGKLESGQ QKISSTTNLS SFPENDLVEL KWQNGQIVMQ GQSSSAKKSP VDNNLPSSAS 60 GDRDKFTGNS STSKIGKFGL MDTMLNDMSL SVPTGELDLI QEDEGVPWLG YSADDSLQQD 120 YCSQLLPEIS GVTANKQSGQ SVFGLINKRG SSDKMIGDSH SVPVHNAASL EQRNTSKVSP 180 SSRFSLLSSL ASQKGHASVP TLESGVADTF SSKKSNTPMS VLGDSSQNQA SAGDVKSDKS 240 QKQNMPGNRS NLLNFSHFSR PATFVKAAKL QNITGGSNVS GSSVLEAKGK KGGEKVDNHV 300 SAAAAENYLP SKKENVLHYP TNAVSSQLES RPSGASFHDR SGQVEQSDNA FRDCSSNNDN 360 HRDQFTGAKA IRDIADGERN VDHGVACSSV CSGSSAERGS SDQPLNLKRK NRDNEESECR 420 SEDVEEESVG IKKTCAARGG TGSKRSRAAE VHNLSERRRR DRINEKMRAL QELIPNCNKA 480 DKASMLDEAI EYLKTLQLQV QIMSMGAGFC VPPMMFPTGV QHMHGAQMPH YSPMSLGIGM 540 GMGMGFGMGM LEMNGRSSGY PIFPMPSVQG GHFPSPPIPA STAYPGIAVS NRHAFAHPGQ 600 GLPMSVPRAS LVPLAGQLST GATVPPNVAR AGVPVEIQSA PPIMDSKNPV HKNSRIVHNV 660 EPSLPQNHTC NQVQATNEVL DRSALKNDQL PDVTDCAANN LTNPTNVPGN ETGEFVLENN 720 NFIRYANK |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 455 | 460 | ERRRRD |
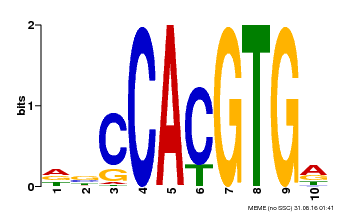
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00081 | ChIP-seq | Transfer from AT1G09530 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | HG975440 | 0.0 | HG975440.1 Solanum pennellii chromosome ch01, complete genome. | |||
| GenBank | HG975513 | 0.0 | HG975513.1 Solanum lycopersicum chromosome ch01, complete genome. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006358519.1 | 0.0 | PREDICTED: transcription factor PIF3-like | ||||
| Refseq | XP_015169448.1 | 0.0 | PREDICTED: transcription factor PIF3-like | ||||
| TrEMBL | M1BK41 | 0.0 | M1BK41_SOLTU; Uncharacterized protein | ||||
| STRING | PGSC0003DMT400047079 | 0.0 | (Solanum tuberosum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA4965 | 24 | 37 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09530.2 | 9e-41 | phytochrome interacting factor 3 | ||||



