 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sme2.5_03789.1_g00003.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Solaneae; Solanum
|
||||||||
| Family | MIKC_MADS | ||||||||
| Protein Properties | Length: 272aa MW: 30688.8 Da PI: 7.4514 | ||||||||
| Description | MIKC_MADS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SRF-TF | 90.1 | 1.1e-28 | 9 | 59 | 1 | 51 |
S---SHHHHHHHHHHHHHHHHHHHHHHHHHHT-EEEEEEE-TTSEEEEEE- CS
SRF-TF 1 krienksnrqvtfskRrngilKKAeELSvLCdaevaviifsstgklyeyss 51
k ien nrqvtfskRr+g+lKKA+ELSvLCd evaviifsstgkl+e+ss
Sme2.5_03789.1_g00003.1 9 KLIENVNNRQVTFSKRRAGLLKKASELSVLCDSEVAVIIFSSTGKLFEFSS 59
569**********************************************96 PP
| |||||||
| 2 | K-box | 56.1 | 1.6e-19 | 105 | 189 | 4 | 88 |
K-box 4 ssgksleeakaeslqqelakLkkeienLqreqRhllGedLesLslkeLqqLeqqLekslkkiRskKnellleqieelqkkekelq 88
+s++++++++ ++ q+e++ Lk+e+++L+++q +llG+dL+ + l+eL+ Le+qL+++l i+++K+ell++q+e+++k+ +el+
Sme2.5_03789.1_g00003.1 105 QSQSQQQTHELKQEQKEVDSLKDELSKLKTKQLRLLGKDLNGMGLNELRLLEHQLNEGLLGIKERKEELLIQQLEYSRKQVEELR 189
4444488889999*******************************************************************99986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50066 | 31.525 | 1 | 61 | IPR002100 | Transcription factor, MADS-box |
| SMART | SM00432 | 3.7E-40 | 1 | 60 | IPR002100 | Transcription factor, MADS-box |
| CDD | cd00265 | 5.70E-36 | 2 | 90 | No hit | No description |
| SuperFamily | SSF55455 | 9.94E-29 | 2 | 84 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 5.0E-28 | 3 | 23 | IPR002100 | Transcription factor, MADS-box |
| PROSITE pattern | PS00350 | 0 | 3 | 57 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF00319 | 1.2E-25 | 11 | 57 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 5.0E-28 | 23 | 38 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 5.0E-28 | 38 | 59 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF01486 | 2.6E-13 | 110 | 188 | IPR002487 | Transcription factor, K-box |
| PROSITE profile | PS51297 | 10.657 | 115 | 201 | IPR002487 | Transcription factor, K-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010047 | Biological Process | fruit dehiscence | ||||
| GO:0010262 | Biological Process | somatic embryogenesis | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0048577 | Biological Process | negative regulation of short-day photoperiodism, flowering | ||||
| GO:0060862 | Biological Process | negative regulation of floral organ abscission | ||||
| GO:0060867 | Biological Process | fruit abscission | ||||
| GO:0071365 | Biological Process | cellular response to auxin stimulus | ||||
| GO:2000692 | Biological Process | negative regulation of seed maturation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005737 | Cellular Component | cytoplasm | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 272 aa Download sequence Send to blast |
MGRGKIDIKL IENVNNRQVT FSKRRAGLLK KASELSVLCD SEVAVIIFSS TGKLFEFSST 60 SLKCNAAQDK IKLSYSMKQT LSRYNRCVAS TETSAAEKKS EDKEQSQSQQ QTHELKQEQK 120 EVDSLKDELS KLKTKQLRLL GKDLNGMGLN ELRLLEHQLN EGLLGIKERK EELLIQQLEY 180 SRKQVEELRG LFPLSASLPP PYLEFHHPME KKYPILKVSE ESLDSDTACE DGVDDEDSNT 240 TLQLGLPTIG RKRKKAEQES PSSNSENQVG SK |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5f28_A | 8e-21 | 1 | 76 | 1 | 70 | MEF2C |
| 5f28_B | 8e-21 | 1 | 76 | 1 | 70 | MEF2C |
| 5f28_C | 8e-21 | 1 | 76 | 1 | 70 | MEF2C |
| 5f28_D | 8e-21 | 1 | 76 | 1 | 70 | MEF2C |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 249 | 255 | GRKRKKA |
| 2 | 250 | 254 | RKRKK |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00508 | DAP | Transfer from AT5G13790 | Download |
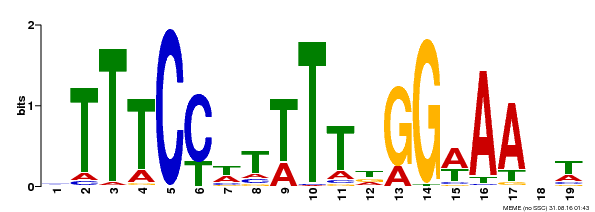
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT014220 | 1e-115 | BT014220.1 Lycopersicon esculentum clone 133400R, mRNA sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_016538893.1 | 1e-160 | PREDICTED: agamous-like MADS-box protein AGL15 | ||||
| TrEMBL | A0A2G2XQH5 | 1e-163 | A0A2G2XQH5_CAPBA; Uncharacterized protein | ||||
| STRING | Solyc01g087990.2.1 | 1e-156 | (Solanum lycopersicum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA8775 | 22 | 26 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G13790.1 | 2e-54 | AGAMOUS-like 15 | ||||



