 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sme2.5_06280.1_g00002.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Solaneae; Solanum
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 475aa MW: 52481.6 Da PI: 6.2139 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 51.7 | 2e-16 | 57 | 101 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r rW++eE++++++a k++G W++I +++g ++t+ q++s+ qk+
Sme2.5_06280.1_g00002.1 57 RERWSEEEHKKFLEALKLHGRA-WRRIEEHVG-TKTAVQIRSHAQKF 101
78******************88.*********.************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 5.08E-16 | 51 | 105 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 21.204 | 52 | 106 | IPR017930 | Myb domain |
| TIGRFAMs | TIGR01557 | 2.9E-15 | 55 | 104 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 5.5E-13 | 56 | 104 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 5.2E-9 | 57 | 97 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 6.8E-14 | 57 | 100 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 3.13E-10 | 59 | 102 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0007623 | Biological Process | circadian rhythm | ||||
| GO:0009734 | Biological Process | auxin-activated signaling pathway | ||||
| GO:0010600 | Biological Process | regulation of auxin biosynthetic process | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 475 aa Download sequence Send to blast |
MAIRDQSESD TILPSRNRIS VDVGAPPPRN VLLEEQVTQE EEFAPKIRKP YTISKQRERW 60 SEEEHKKFLE ALKLHGRAWR RIEEHVGTKT AVQIRSHAQK FFSKVVRESS NGDASCVKSI 120 EIPPPRPKRK PMHPYPRKMA TPLTNGTLVP EKLKRSASPD LSLSEPENQS PTSVLSTLGS 180 DAFGTVDSTK PIENSSPVSS VVAENSGDLL LSEPSGFILE ERRSSPAQAY ASSNPDNQTC 240 MKLDLFPEEN DFVKEGSDEA SSTQCLKLFG KTVLVTDTHM TSSTSGQISL TDANDDASRT 300 LPRSSFPTKY LPWDLECLQS TFTVGPPTRS YYFPAPNGSS GPNQSVSSPL LSWGSSYASP 360 CTQVHNPIPI KRRLLFSDID LEDKETQKEG SSTSSNTESV DAELSGDKNL EIEAQSSSRN 420 VLEESVRASL PSVLANSMKR VKGFVPYKRC LTERGISSST LTGEEREEQR TRLCL |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Morning-phased transcription factor integrating the circadian clock and auxin pathways. Binds to the evening element (EE) of promoters. Does not act within the central clock, but regulates free auxin levels in a time-of-day specific manner. Positively regulates the expression of YUC8 during the day, but has no effect during the night. Negative regulator of freezing tolerance. {ECO:0000269|PubMed:19805390, ECO:0000269|PubMed:23240770}. | |||||
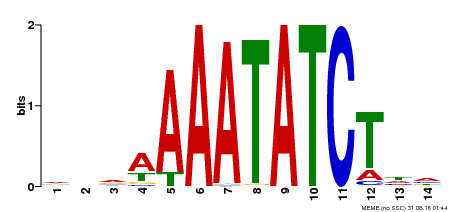
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00515 | DAP | Transfer from AT5G17300 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Circadian-regulation. Peak of transcript abundance near subjective dawn. Down-regulated and strongly decreased amplitude of circadian oscillation upon cold treatment. {ECO:0000269|PubMed:19805390, ECO:0000269|PubMed:23240770}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006350462.1 | 0.0 | PREDICTED: protein REVEILLE 1 | ||||
| Swissprot | F4KGY6 | 4e-61 | RVE1_ARATH; Protein REVEILLE 1 | ||||
| TrEMBL | M1BXP3 | 0.0 | M1BXP3_SOLTU; Uncharacterized protein | ||||
| STRING | PGSC0003DMT400055263 | 0.0 | (Solanum tuberosum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA3185 | 24 | 47 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G17300.1 | 2e-56 | MYB_related family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



