 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sobic.001G002500.1.p | ||||||||
| Common Name | Sb01g000352, SORBIDRAFT_01g000352 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Andropogonodae; Andropogoneae; Sorghinae; Sorghum
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 295aa MW: 30988.6 Da PI: 8.2762 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 58.1 | 2.1e-18 | 151 | 202 | 2 | 56 |
AP2 2 gykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkklege 56
y+GVr ++ +g+++AeIrd ++ng r++lg+f++ae Aa a+++a+ +++g+
Sobic.001G002500.1.p 151 PYRGVRKRP-WGKFAAEIRDSTSNG--VRVWLGTFDSAEAAALAYDQAAFAMRGA 202
59*******.**********66687..*************************995 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00380 | 1.8E-36 | 151 | 215 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 8.0E-30 | 151 | 210 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 22.813 | 151 | 209 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 7.37E-14 | 152 | 186 | No hit | No description |
| SuperFamily | SSF54171 | 1.05E-21 | 152 | 211 | IPR016177 | DNA-binding domain |
| PRINTS | PR00367 | 5.3E-10 | 152 | 163 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 7.7E-13 | 152 | 201 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 5.3E-10 | 175 | 191 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0006952 | Biological Process | defense response | ||||
| GO:0009873 | Biological Process | ethylene-activated signaling pathway | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 295 aa Download sequence Send to blast |
MVSSTSHSQN SNNSPPPPPP VHLSTGIVGF LARRRAAMAT THHPSSFQFQ QPRVAAAPLD 60 SPGSSTSSVD SSAPWSCMAQ AQQQHRAPPA PVLPFDSNDT DEMALLDMIA GQGQEEAGLV 120 LATSKQEEAE ANNGTGGAVG PGAGAGAGGR PYRGVRKRPW GKFAAEIRDS TSNGVRVWLG 180 TFDSAEAAAL AYDQAAFAMR GAAAVLNFPV ERVRQSLEGM GMGTGTDSGS PVVALKRRHS 240 MRMRVRRPAS RSRKAKAATR SEVMELEDLG ADYLEALLGA TEESESLCRT HHSI* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5wx9_A | 1e-27 | 142 | 211 | 5 | 74 | Ethylene-responsive transcription factor ERF096 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to the GCC-box cis-acting elements found in the promoter regions of ethylene-responsive genes (PubMed:23057995). Acts downstream of MYC2 in the jasmonate-mediated response to Botrytis cinerea infection (PubMed:28733419). With MYC2 forms a transcription module that regulates pathogen-responsive genes (PubMed:28733419). {ECO:0000269|PubMed:23057995, ECO:0000269|PubMed:28733419}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00374 | DAP | Transfer from AT3G23240 | Download |
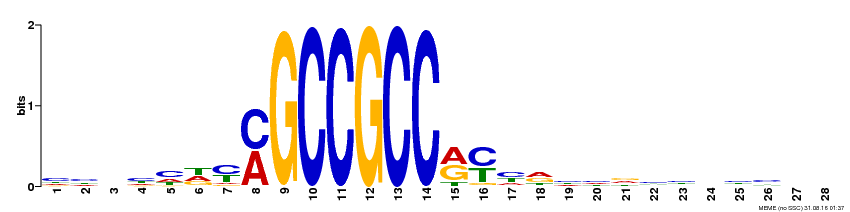
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Sobic.001G002500.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by wounding and infection with the fungal pathogen Botrytis cinerea. {ECO:0000269|PubMed:28733419}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT069980 | 1e-94 | BT069980.1 Zea mays full-length cDNA clone ZM_BFb0349F03 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002463464.2 | 0.0 | ethylene-responsive transcription factor 1B | ||||
| Swissprot | A0A3Q7I5Y9 | 5e-54 | ERFC3_SOLLC; Ethylene-response factor C3 | ||||
| TrEMBL | C5WRV0 | 0.0 | C5WRV0_SORBI; Uncharacterized protein | ||||
| STRING | Sb01g000352.1 | 0.0 | (Sorghum bicolor) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP277 | 37 | 249 | Representative plant | OGRP6 | 16 | 1718 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G23240.1 | 4e-27 | ethylene response factor 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Sobic.001G002500.1.p |
| Entrez Gene | 8059234 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



