 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sobic.001G026500.1.p | ||||||||
| Common Name | Sb01g002530, SORBIDRAFT_01g002530 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Andropogonodae; Andropogoneae; Sorghinae; Sorghum
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 970aa MW: 105843 Da PI: 5.1301 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 131.4 | 3.2e-41 | 151 | 228 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+CqvegC adl+ ak+yhrrhkvCe+h+ka++++v ++ qrfCqqCsrfh l+efDe+krsCrrrLa+hn+rrrk+++
Sobic.001G026500.1.p 151 ACQVEGCGADLTAAKDYHRRHKVCEMHAKASTAVVGNTVQRFCQQCSRFHLLQEFDEGKRSCRRRLAGHNRRRRKTRP 228
5**************************************************************************875 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 2.1E-34 | 144 | 213 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 32.624 | 149 | 226 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 6.54E-39 | 150 | 230 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 4.6E-30 | 152 | 225 | IPR004333 | Transcription factor, SBP-box |
| CDD | cd00204 | 1.49E-9 | 726 | 856 | No hit | No description |
| SMART | SM00248 | 0.24 | 751 | 780 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50297 | 9.145 | 751 | 818 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 2.64E-8 | 752 | 857 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 3.2E-8 | 753 | 856 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 460 | 797 | 827 | IPR002110 | Ankyrin repeat |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0016021 | Cellular Component | integral component of membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 970 aa Download sequence Send to blast |
MEAARFGTQS SHLYGGGLGE LDLNRRENRV FGWDLNDWSW DSQRFVATPV PAQAANGSGL 60 NSSPSSSEEA EEDVARNGGL RGDTDKRKRV VVIDDDETED QDTVVNGGGS LSLRIGGTAV 120 GVEAMENSDV NEDERNGKKI RVQGGGSSGP ACQVEGCGAD LTAAKDYHRR HKVCEMHAKA 180 STAVVGNTVQ RFCQQCSRFH LLQEFDEGKR SCRRRLAGHN RRRRKTRPDT AVGGTASIED 240 KVSNYLLLSL IGICANLNSD NVQHSNSQEL LSTLLKNLGS VAKSLEPKEL CKLLEAYQSL 300 QNGSNAGTSG TANVAEEAAG PSNSKLPFVN GSHCGQASSS VVPVQSKATM VVTPEPASCK 360 LKDFDLNDTC NDMEGFEDGQ EGSPTPAFKT ADSPNCASWM QQDSTQSPPQ TSGNSDSTST 420 QSLSSSNGDT QCRTDKIVFK LFDKVPGDLP PVLRSQILGW LSSSPTDIES HIRPGCIILT 480 VYLRLVESAW QELSENMSLH LDKLLSSSTD DFWASGLVFV MVRRHLAFML NGQIMLDRPL 540 APSSHDYCKI LCVKPVAAPY SATINFRVEG FNLLSTSSRL ICSFEGRCIF QEDTDFIAEN 600 AECEDRDIEC LSFCCSVPGP RGRGFIEVED SGFSNGFFPF IIAEKDVCSE VSELESIFES 660 SSNEHANVND DARDQALEFI NELGWLLHRA NRMSKEDETD TSLAALNMWR FRNLGVFAME 720 REWCAVIKML LDFLFIGLVD VGSRSPEDVV LSENLLHAAV RRKSVNMVRF LLRYKPNKNS 780 KGTAQTYLFR PDALGPSTIT PLHIAAATSD AEDVLDVLTD DPGLIGISAW SNARDETGFT 840 PEDYARQRGN DAYLNLVQKK IDKHLGKGHV VLGVPSSICS VITDGVKPGD VSLEICRPMS 900 AAVPSCLLCS RQARVYPNST TKTFLYRPAM LTVMGVAVVC VCVGILLHTF PRVYAAPTFR 960 WESLERGAM* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 4e-31 | 146 | 225 | 1 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Sbi.3741 | 0.0 | leaf| panicle| shoot | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Ubiquitous. {ECO:0000269|PubMed:16861571}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00633 | PBM | Transfer from PK06791.1 | Download |
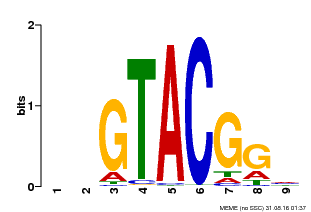
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Sobic.001G026500.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT038852 | 0.0 | BT038852.1 Zea mays full-length cDNA clone ZM_BFb0336G12 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002466159.1 | 0.0 | squamosa promoter-binding-like protein 6 | ||||
| Swissprot | Q75LH6 | 0.0 | SPL6_ORYSJ; Squamosa promoter-binding-like protein 6 | ||||
| TrEMBL | C5WU18 | 0.0 | C5WU18_SORBI; Uncharacterized protein | ||||
| STRING | Sb01g002530.1 | 0.0 | (Sorghum bicolor) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1998 | 38 | 92 | Representative plant | OGRP2595 | 14 | 32 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G60030.1 | 0.0 | squamosa promoter-binding protein-like 12 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Sobic.001G026500.1.p |
| Entrez Gene | 8079685 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



