 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sobic.001G120900.1.p | ||||||||
| Common Name | Sb01g010660, SORBIDRAFT_01g010660 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Andropogonodae; Andropogoneae; Sorghinae; Sorghum
|
||||||||
| Family | GRAS | ||||||||
| Protein Properties | Length: 628aa MW: 66272.3 Da PI: 4.8982 | ||||||||
| Description | GRAS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GRAS | 459.8 | 1.7e-140 | 245 | 623 | 1 | 374 |
GRAS 1 lvelLlecAeavssgdlelaqalLarlselaspdgdpmqRlaayfteALaarlarsvselykalppsetseknsseelaalklfsevsP 89
lv++Ll+cAeav+++++++a+al++++ las++g +m+++aayf eALa+r++r ++ p ++ + +++ l+a f+e +P
Sobic.001G120900.1.p 245 LVHALLACAEAVQQENFSAADALVKQIPMLASSQGGAMRKVAAYFGEALARRVYR-----FRPTPDTSLLDAAVADFLHA--HFYESCP 326
689****************************************************.....43333333333445555555..4****** PP
GRAS 90 ilkfshltaNqaIleavegeervHiiDfdisqGlQWpaLlqaLasRpegppslRiTgvgspesgskeeleetgerLakfAeelgvpfef 178
+lkf+h+taNqaIlea++g++rvH++Df+i+qGlQWpaLlqaLa Rp+gpps+R+Tgvg+p+++++++l+++g++La+fA++++v+f++
Sobic.001G120900.1.p 327 YLKFAHFTANQAILEAFAGCRRVHVVDFGIKQGLQWPALLQALALRPGGPPSFRLTGVGPPQHDETDALQQVGWKLAQFAHTIRVDFQY 415
***************************************************************************************** PP
GRAS 179 nvlvakrledleleeLrvkp......gEalaVnlvlqlhrlldesvsleserdevLklvkslsPkvvvvveqeadhnsesFlerfleal 261
+ lva +l+dle+ +L+ + E++aVn+v++lhrll+++++le+ vL +v+ ++P++v+vveqea+hns++Fl+rf+e+l
Sobic.001G120900.1.p 416 RGLVAATLADLEPFMLQPEGddkdeePEVIAVNSVFELHRLLAQPGALEK----VLGTVRAVRPRIVTVVEQEANHNSGTFLDRFTESL 500
****************65544555679***********************....*********************************** PP
GRAS 262 eyysalfdsleak........lpres.eerikvErellgreivnvvacegaerrerhetlekWrerleeaGFkpvplsekaakqaklll 341
+yys++fdsle p+++ +++++++++lgr+i+nvvacegaer+erhetl++Wr rl +GF+pv+l+++a+kqa++ll
Sobic.001G120900.1.p 501 HYYSTMFDSLEGAgsgqstdaSPAAAgGTDQVMSEVYLGRQICNVVACEGAERTERHETLSQWRGRLVGSGFEPVHLGSNAYKQASTLL 589
**********7775777766544444799************************************************************ PP
GRAS 342 rkvk.sdgyrveeesgslvlgWkdrpLvsvSaWr 374
+ ++ +dgyrvee++g+l+lgW++rpL+++SaWr
Sobic.001G120900.1.p 590 ALFNgGDGYRVEEKDGCLTLGWHTRPLIATSAWR 623
*********************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF12041 | 6.3E-35 | 37 | 113 | IPR021914 | Transcriptional factor DELLA, N-terminal |
| SMART | SM01129 | 3.1E-31 | 37 | 119 | No hit | No description |
| PROSITE profile | PS50985 | 67.736 | 219 | 602 | IPR005202 | Transcription factor GRAS |
| Pfam | PF03514 | 5.8E-138 | 245 | 623 | IPR005202 | Transcription factor GRAS |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009863 | Biological Process | salicylic acid mediated signaling pathway | ||||
| GO:0009867 | Biological Process | jasmonic acid mediated signaling pathway | ||||
| GO:0009938 | Biological Process | negative regulation of gibberellic acid mediated signaling pathway | ||||
| GO:0010187 | Biological Process | negative regulation of seed germination | ||||
| GO:0010218 | Biological Process | response to far red light | ||||
| GO:0042176 | Biological Process | regulation of protein catabolic process | ||||
| GO:0042538 | Biological Process | hyperosmotic salinity response | ||||
| GO:2000033 | Biological Process | regulation of seed dormancy process | ||||
| GO:2000377 | Biological Process | regulation of reactive oxygen species metabolic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 628 aa Download sequence Send to blast |
MKREYQDAGG SGGDMGSSKD KMMVAAAGAG EQEEELDEML ASLGYKVRSS DMADVAQKLE 60 QLEMAMGMGG VGGAGATADD GFISHLATDT VHYNPSDLSS WLESMLSELN APPPPLPPAT 120 TPPAPRLAST SSTVTSGAAA GAGYFDLPPA VDSSSSTYAL KPIPSPVAVA SADPSSTDST 180 REPKRMRTGG GSTSSSSSSS SSMDGGRTRS SVVEAAPPAT QAPAAANGPA VPVVVMDTQE 240 AGIRLVHALL ACAEAVQQEN FSAADALVKQ IPMLASSQGG AMRKVAAYFG EALARRVYRF 300 RPTPDTSLLD AAVADFLHAH FYESCPYLKF AHFTANQAIL EAFAGCRRVH VVDFGIKQGL 360 QWPALLQALA LRPGGPPSFR LTGVGPPQHD ETDALQQVGW KLAQFAHTIR VDFQYRGLVA 420 ATLADLEPFM LQPEGDDKDE EPEVIAVNSV FELHRLLAQP GALEKVLGTV RAVRPRIVTV 480 VEQEANHNSG TFLDRFTESL HYYSTMFDSL EGAGSGQSTD ASPAAAGGTD QVMSEVYLGR 540 QICNVVACEG AERTERHETL SQWRGRLVGS GFEPVHLGSN AYKQASTLLA LFNGGDGYRV 600 EEKDGCLTLG WHTRPLIATS AWRLAAP* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5b3h_A | 4e-61 | 228 | 622 | 1 | 377 | Protein SCARECROW |
| 5b3h_D | 4e-61 | 228 | 622 | 1 | 377 | Protein SCARECROW |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Sbi.9966 | 0.0 | callus| embryo| panicle| root | ||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcriptional regulator that acts as a repressor of the gibberellin (GA) signaling pathway. Probably acts by participating in large multiprotein complexes that repress transcription of GA-inducible genes. Upon GA application, it is degraded by the proteasome, allowing the GA signaling pathway. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00611 | ChIP-seq | Transfer from AT2G01570 | Download |
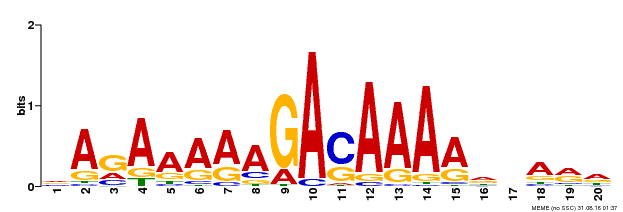
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Sobic.001G120900.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AJ242530 | 0.0 | AJ242530.1 Zea mays partial d8 gene for gibberellin response modulator. | |||
| GenBank | BT054262 | 0.0 | BT054262.1 Zea mays full-length cDNA clone ZM_BFb0203D23 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002466594.1 | 0.0 | DELLA protein DWARF8 | ||||
| Swissprot | Q9ST48 | 0.0 | DWRF8_MAIZE; DELLA protein DWARF8 | ||||
| TrEMBL | C5WNR2 | 0.0 | C5WNR2_SORBI; Uncharacterized protein | ||||
| STRING | Sb01g010660.1 | 0.0 | (Sorghum bicolor) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP998 | 38 | 138 | Representative plant | OGRP1251 | 15 | 50 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G14920.1 | 1e-171 | GRAS family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Sobic.001G120900.1.p |
| Entrez Gene | 8062929 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



