 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sobic.001G261501.2.p | ||||||||
| Common Name | Sb0012s010230, SORBIDRAFT_0012s010230, SORBI_K006800 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Andropogonodae; Andropogoneae; Sorghinae; Sorghum
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 1022aa MW: 113463 Da PI: 5.7916 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 172 | 8.6e-54 | 21 | 138 | 2 | 118 |
CG-1 2 lke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahse 89
lke ++rwl++ ei++iL+n+++++++ e+++rp+sgsl+L++rk++ryfrkDG++w+kk d ktv+E+he+LK g+++vl+cyYah+e
Sobic.001G261501.2.p 21 LKEaQHRWLRPAEICEILKNYRNFRIAPEPPNRPPSGSLFLFDRKVLRYFRKDGHNWRKKNDQKTVKEAHERLKSGSIDVLHCYYAHGE 109
45559************************************************************************************ PP
CG-1 90 enptfqrrcywlLeeelekivlvhylevk 118
en +fqrr+yw+Lee++ +ivlvhyle+k
Sobic.001G261501.2.p 110 ENINFQRRTYWMLEEDYMHIVLVHYLETK 138
**************************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 80.861 | 17 | 143 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 2.7E-72 | 20 | 138 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 6.8E-48 | 23 | 136 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:2.60.40.10 | 2.7E-4 | 445 | 532 | IPR013783 | Immunoglobulin-like fold |
| SuperFamily | SSF81296 | 7.4E-16 | 446 | 532 | IPR014756 | Immunoglobulin E-set |
| Pfam | PF01833 | 8.0E-6 | 446 | 524 | IPR002909 | IPT domain |
| CDD | cd00204 | 1.73E-13 | 591 | 738 | No hit | No description |
| SuperFamily | SSF48403 | 8.08E-17 | 614 | 740 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 4.3E-17 | 641 | 741 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50088 | 8.683 | 647 | 679 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50297 | 17.29 | 647 | 740 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50088 | 8.736 | 680 | 712 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 1.78E-8 | 839 | 892 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 0.086 | 841 | 863 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.407 | 842 | 871 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.0032 | 843 | 862 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0022 | 864 | 886 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.56 | 865 | 889 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 2.0E-4 | 867 | 886 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0045944 | Biological Process | positive regulation of transcription from RNA polymerase II promoter | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0001077 | Molecular Function | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1022 aa Download sequence Send to blast |
MASAEARRLA VVPQLEIEQI LKEAQHRWLR PAEICEILKN YRNFRIAPEP PNRPPSGSLF 60 LFDRKVLRYF RKDGHNWRKK NDQKTVKEAH ERLKSGSIDV LHCYYAHGEE NINFQRRTYW 120 MLEEDYMHIV LVHYLETKGG KSSRARGNNN IIQGTAVDSP VSQLPSQTME GESSLSGQAS 180 EYEEAESDIY SGAGYNSFTW MQQHENGTGP VTNSSVFSSY TPASSVGNYQ GLHATQNTSF 240 YPVNQHNSPL ILNGSSAMLG TNGRANQTDL PSWNSVIELD EPVQMPHLQF PVPPDQSATT 300 EGLGVDYLTF DEVYSDGLSL KDIGAAGTHG ESYLQFSSAT GDLSATENSF PQQNDGSLEA 360 AIGYPFLKTQ SSNLSDILKD SFKKTDSFTR WMSKELPEVE DSQIHSSSGG FWSTEEANNI 420 IEASSREPLD QFTVSPMLSQ DQLFSIVDFA PNWTYVGSKT KILVAGSILN DSQINEGCKW 480 SCMFGEVEVP AKVLADGTLI CYSPQHRPGR VPFYITCSNR LACSEVREFE FRPTVSQYMD 540 APSPHGETNK VYFQIRLDKL LSLGPDEYQA TVSNPSLEMI DLSKKISSLM ASNDEWSNLL 600 KLAVDNEPST ADQHDQFAEN LIKEKLHVWL LNKVGMGGKG PSVLDDEGQG VLHLAAALGY 660 DWAIRPTLAA GVNINFRDVH GWTALHWAAF CGRERTVVAL IALGAAPGAL TDPTPDFPGS 720 TPADLASANG QKGISGFLAE SSLTSHLQAL NLKEANMSQI SGLPGIGDVT ERDSLQPPSG 780 DSLGPVRNAA QAAAQIYQVF RVQSFQRKQA AQYEDDKGGM SDERALSLLS VKPPKSGQLD 840 PLHSAATRIQ NKFRGWKGRK EFLLIRQRIV KIQAHVRGHQ VRKHYRKIVW SVGIVEKVIL 900 RWRRRGAGLR GFRSTEGSVE SSSGGTSSSS IQDKPSGDDY DFLQEGRKQT EERLQKALAR 960 VKSMAQYPEA RDQYQRILTV VSKMQESQAM QEKMLEESAE MDFMSEFKEL WDDDTPIPGY 1020 F* |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Sbi.5793 | 0.0 | embryo| leaf| ovary| panicle| pollen | ||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00042 | PBM | Transfer from AT2G22300 | Download |
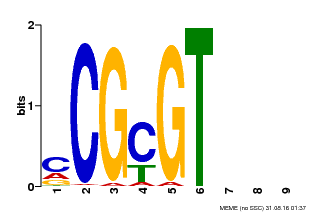
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Sobic.001G261501.2.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002489212.1 | 0.0 | calmodulin-binding transcription activator 3 isoform X2 | ||||
| TrEMBL | C6JRU1 | 0.0 | C6JRU1_SORBI; Uncharacterized protein | ||||
| STRING | Sb0012s010230.1 | 0.0 | (Sorghum bicolor) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G64220.2 | 0.0 | Calmodulin-binding transcription activator protein with CG-1 and Ankyrin domains | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Sobic.001G261501.2.p |
| Entrez Gene | 8155742 |



