 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sobic.002G247500.4.p | ||||||||
| Common Name | Sb02g028390, SORBIDRAFT_02g028390 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Andropogonodae; Andropogoneae; Sorghinae; Sorghum
|
||||||||
| Family | EIL | ||||||||
| Protein Properties | Length: 610aa MW: 68172.7 Da PI: 6.1838 | ||||||||
| Description | EIL family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | EIN3 | 422.6 | 4.2e-129 | 38 | 414 | 1 | 351 |
XXXXXXXXXXXXXXXXXXXXXXX..XXXXX.XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX CS
EIN3 1 eelkkrmwkdqmllkrlkerkkqlledkeaatgakksnksneqarrkkmsraQDgiLkYMlkemevcnaqGfvYgiipekgkpvegasd 89
e+l +rmwkd+++l+r+ker+++l ++a++++ + + +++qa rkkmsra DgiLkYMlk m+vcna+GfvYgiip+kgkpv+gasd
Sobic.002G247500.4.p 38 EDLARRMWKDRVRLRRIKERQQKLA-LQQAELEKLRPKPISDQAMRKKMSRAHDGILKYMLKLMQVCNARGFVYGIIPDKGKPVSGASD 125
89*******************9865.66667********************************************************** PP
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX....XX----STTS-HHHHHHHHHHHSSSSSS-TTS--TTT--HHHH---S-- CS
EIN3 90 sLraWWkekvefdrngpaaiskyqaknlilsgesslqtersseshslselqDTtlgSLLsalmqhcdppqrrfplekgvepPWWPtGke 178
++raWWkekv+fd+ngpaai ky+++nl+ ++++s +s+++hsl++lqD+tlgSLLs+lmqhcdppqr++plekgv+pPWWP+Gke
Sobic.002G247500.4.p 126 NIRAWWKEKVKFDKNGPAAIEKYDSENLVTANAQSG---GSKNPHSLMDLQDATLGSLLSSLMQHCDPPQRKYPLEKGVPPPWWPSGKE 211
******************************988888...9************************************************* PP
HHHHHHT--TT--.-----GGG--HHHHHHHHHHHHHHTGGGHHHHHHTTTTSSSSTTT--SHHHHHHHHHHTTTTT-S--XXXX.XX. CS
EIN3 179 lwwgelglskdqgtppykkphdlkkawkvsvLtavikhmsptieeirelerqskylqdkmsakesfallsvlnqeekecatvsah.ss. 265
+ww +lgl+ q ppykkphdlkk+wk +vLt vikhmsp++++ir+++r+sk+lqdkm+akes ++l vl++ee++++++ s+
Sobic.002G247500.4.p 212 EWWIALGLPSGQI-PPYKKPHDLKKVWKAGVLTGVIKHMSPNFDKIRNHVRKSKCLQDKMTAKESLIWLGVLQREESLVHRIDNGvSEi 299
***********99.9******************************************************************99986335 PP
...XXXXXXXXXXXXXXXXXXXXXXXXXX.XXXXXXXXXX...............................XXXXXXXXXXXXXXXXXX CS
EIN3 266 ...slrkqspkvtlsceqkedvegkkeskikhvqavktta...............................gfpvvrkrkkkpsesakv 320
s+ ++++ t s+++++dv g ++ +++++++++++ +++ krkk +++s+ +
Sobic.002G247500.4.p 300 tprSMPEDRNGDTNSSSNEYDVYGFEDAPVSTSSKDDEQDlspvtqsavehvpkrgreraynkhpnqivpgKAKEPPKRKKARRSSTVT 388
55588889999999*********999999999999999889999999999999999999999999777776666666666444444444 PP
XXX......XXXXXXX.XXXXXXXXXXXXXX CS
EIN3 321 sskevsrtcqssqfrgsetelifadknsisq 351
+ + + +++++ ++d+n+++q
Sobic.002G247500.4.p 389 EPDA----RRVVDAP-ENSRNLIPDMNRLDQ 414
4332....2333333.356666777777666 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04873 | 3.9E-124 | 38 | 286 | No hit | No description |
| Gene3D | G3DSA:1.10.3180.10 | 1.7E-69 | 161 | 293 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| SuperFamily | SSF116768 | 5.89E-60 | 166 | 289 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0042762 | Biological Process | regulation of sulfur metabolic process | ||||
| GO:0071281 | Biological Process | cellular response to iron ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 610 aa Download sequence Send to blast |
MDQLAMLATE LGDSSDFEVE GIQNLTENDV SDEEIEPEDL ARRMWKDRVR LRRIKERQQK 60 LALQQAELEK LRPKPISDQA MRKKMSRAHD GILKYMLKLM QVCNARGFVY GIIPDKGKPV 120 SGASDNIRAW WKEKVKFDKN GPAAIEKYDS ENLVTANAQS GGSKNPHSLM DLQDATLGSL 180 LSSLMQHCDP PQRKYPLEKG VPPPWWPSGK EEWWIALGLP SGQIPPYKKP HDLKKVWKAG 240 VLTGVIKHMS PNFDKIRNHV RKSKCLQDKM TAKESLIWLG VLQREESLVH RIDNGVSEIT 300 PRSMPEDRNG DTNSSSNEYD VYGFEDAPVS TSSKDDEQDL SPVTQSAVEH VPKRGRERAY 360 NKHPNQIVPG KAKEPPKRKK ARRSSTVTEP DARRVVDAPE NSRNLIPDMN RLDQVEIQGR 420 SNQIVIFDHG DTTTEALQHR GDAQVQVHLP GAEVNNFNSA PTANPTPISI CMGDQPLPYQ 480 NNDSTRSRSE NSFPVDAHPG LNNLPSSYQN LPPKQSLPLS MMDHHVVPMG IRAPTDSIPY 540 GDHILGGGSS TSVPGDMQQL IDFPFYGDQD KFVGSSFEGL PLDYISISSP IPDIDDLLHD 600 DDLMEYLGT* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wij_A | 3e-64 | 169 | 291 | 12 | 134 | ETHYLENE-INSENSITIVE3-like 3 protein |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Sbi.4357 | 0.0 | callus | ||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that may be involved in the ethylene response pathway. {ECO:0000269|PubMed:9215635, ECO:0000269|PubMed:9851977}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00649 | PBM | Transfer from LOC_Os09g31400 | Download |
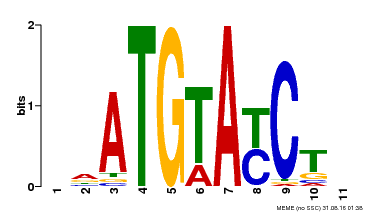
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Sobic.002G247500.4.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT062636 | 0.0 | BT062636.1 Zea mays full-length cDNA clone ZM_BFb0325I05 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002462568.1 | 0.0 | ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| Refseq | XP_021308896.1 | 0.0 | ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| Refseq | XP_021308897.1 | 0.0 | ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| Swissprot | O23116 | 1e-142 | EIL3_ARATH; ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| TrEMBL | C5X3X3 | 0.0 | C5X3X3_SORBI; Uncharacterized protein | ||||
| STRING | Sb02g028390.1 | 0.0 | (Sorghum bicolor) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G73730.1 | 1e-139 | ETHYLENE-INSENSITIVE3-like 3 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Sobic.002G247500.4.p |
| Entrez Gene | 8064849 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



