 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sobic.003G000600.2.p | ||||||||
| Common Name | Sb03g000240, SORBIDRAFT_03g000240 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Andropogonodae; Andropogoneae; Sorghinae; Sorghum
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 435aa MW: 45349.6 Da PI: 9.0545 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 97.6 | 8.1e-31 | 149 | 207 | 2 | 59 |
--SS-EEEEEEE--TT-SS-EEEEEE-ST.T---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsa.gCpvkkkversaedpkvveitYegeHnhe 59
Dg++WrKYGqK+ kg+++pr+YYrCt+a gCpv+k+v+r+a+d +++++tYeg+Hnh+
Sobic.003G000600.2.p 149 ADGCQWRKYGQKMAKGNPCPRAYYRCTMAnGCPVRKQVQRCADDRSILITTYEGTHNHP 207
7***************************999***************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 5.0E-34 | 133 | 209 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 3.27E-28 | 141 | 209 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 29.551 | 143 | 209 | IPR003657 | WRKY domain |
| SMART | SM00774 | 5.4E-37 | 148 | 208 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 1.2E-27 | 150 | 207 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0016036 | Biological Process | cellular response to phosphate starvation | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0080169 | Biological Process | cellular response to boron-containing substance deprivation | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 435 aa Download sequence Send to blast |
MQAELARMND ENQRLRGMLT QVTSSYQALQ MHLVALMQAR AGGQAQLMLP PVAQALPPTT 60 DGAAAAVMPL PRQFLGLGPA AAAEETSNSS TEVGSPRRSS STGGNRRAER GDSPDASTRQ 120 QQVAQQQQEA SMRKARVSVR ARSEAPIIAD GCQWRKYGQK MAKGNPCPRA YYRCTMANGC 180 PVRKQVQRCA DDRSILITTY EGTHNHPLPP AAMAMASTTS AAASMLLSGS MPSGDMMTSN 240 FLARAVLPCS SSMATISASA PFPTVTLDLT HGPPAAARPQ PHFQVPLPPH QQVQQQHHHL 300 QAAALYNAHQ SSSKFSGLHM SSSSTSDNNN NVGTSSRAAV AAADAPPHMD TVTAAAAAIT 360 ADPNFTVALA AAITSIIGGG GGHPIPIAIH HGQGQGQEQG QQQAPTSNSN ANNNNNAVVT 420 SSSNNTATSN SETQ* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2ayd_A | 3e-22 | 135 | 211 | 1 | 76 | WRKY transcription factor 1 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Sbi.19928 | 0.0 | root | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Roots, leaves, shoots, flowers and siliques. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in the control of processes related to senescence and pathogen defense (PubMed:12000796). Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element (PubMed:12000796). Activates the transcription of the SIRK gene and represses its own expression and that of the WRKY42 genes (PubMed:12000796). Modulates phosphate homeostasis and Pi translocation by regulating PHO1 expression (PubMed:25733771). {ECO:0000269|PubMed:12000796, ECO:0000269|PubMed:25733771}. | |||||
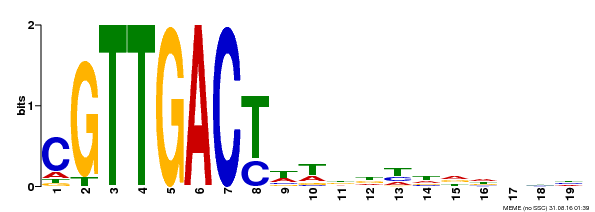
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00209 | DAP | Transfer from AT1G62300 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Sobic.003G000600.2.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By salicylic acid, ethylene, jasmonic acid, pathogens, wounding and strongly during leaf senescence. {ECO:0000269|PubMed:11449049, ECO:0000269|PubMed:11722756}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT085752 | 0.0 | BT085752.2 Zea mays full-length cDNA clone ZM_BFc0057C03 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002457035.1 | 0.0 | WRKY transcription factor 6 isoform X1 | ||||
| Refseq | XP_021313642.1 | 0.0 | WRKY transcription factor 6 isoform X2 | ||||
| Swissprot | Q9C519 | 1e-110 | WRKY6_ARATH; WRKY transcription factor 6 | ||||
| TrEMBL | C5XJG7 | 0.0 | C5XJG7_SORBI; Uncharacterized protein | ||||
| STRING | Sb03g000240.1 | 0.0 | (Sorghum bicolor) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G62300.1 | 6e-81 | WRKY family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Sobic.003G000600.2.p |
| Entrez Gene | 8078066 |



