 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sobic.003G012800.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Andropogonodae; Andropogoneae; Sorghinae; Sorghum
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 393aa MW: 41608.9 Da PI: 4.3228 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 61.8 | 1.5e-19 | 134 | 185 | 1 | 56 |
AP2 1 sgykGVrwdkkrgrWvAeIrd.psengkr.krfslgkfgtaeeAakaaiaarkklege 56
++++GVr+++ +g+++AeIrd + r r++lg+f+taeeAak ++ a+ +l+g+
Sobic.003G012800.1.p 134 PRFRGVRRRP-WGKYAAEIRDpW-----RrVRVWLGTFDTAEEAAKVYDSAAIQLRGA 185
689*******.**********88.....55*************************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF00847 | 4.8E-12 | 134 | 184 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 2.83E-28 | 134 | 194 | No hit | No description |
| SuperFamily | SSF54171 | 1.7E-20 | 135 | 192 | IPR016177 | DNA-binding domain |
| PROSITE profile | PS51032 | 23.196 | 135 | 192 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 9.5E-29 | 135 | 193 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 1.1E-32 | 135 | 198 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 5.4E-10 | 136 | 147 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 5.4E-10 | 158 | 174 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0042991 | Biological Process | transcription factor import into nucleus | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 393 aa Download sequence Send to blast |
MWPMEEEMFV AVRRTEHVEV TSRAVEVAPA AAKGREAAAG SAVPGPRTVR VFCDDYDATD 60 SSSDEAEEDA VARRRVKRYV QEIRLERAVK EEAPAGRAGA SSAAAMAGAA GERTTTRLVL 120 PGRKRKSDGA AAEPRFRGVR RRPWGKYAAE IRDPWRRVRV WLGTFDTAEE AAKVYDSAAI 180 QLRGADATTN FEQVDDAPVP VPDEVAERLP QPPATASAPA PAASKNASSS ATSYDSGEES 240 HAAAASPTSV LRSFPPSAVA DDTCTKAAAK KPTTSPAPAI PAASAPETDE STGGGTSSVF 300 GCPFSSADDC FGGEFPPLYT DFDLLADFPE PSLDFLADIP EEPLSLAPSI PAGSPEEWPS 360 DAEPASPGRW QQVDDFFQDI TDLFQIDPLP VV* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5wx9_A | 8e-19 | 130 | 191 | 9 | 71 | Ethylene-responsive transcription factor ERF096 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00456 | DAP | Transfer from AT4G27950 | Download |
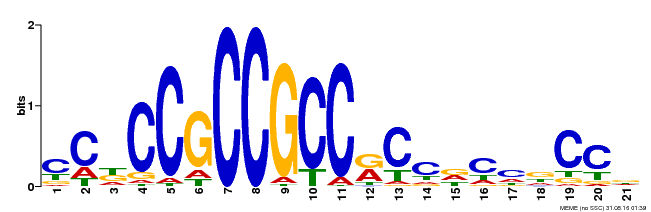
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Sobic.003G012800.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | EU942421 | 4e-89 | EU942421.1 Zea mays clone 1496106 mRNA sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002457105.2 | 0.0 | ethylene-responsive transcription factor CRF2 | ||||
| TrEMBL | A0A1B6Q0S9 | 0.0 | A0A1B6Q0S9_SORBI; Uncharacterized protein | ||||
| STRING | Sb03g001296.1 | 0.0 | (Sorghum bicolor) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1690 | 36 | 97 | Representative plant | OGRP6 | 16 | 1718 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G27950.1 | 5e-32 | cytokinin response factor 4 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Sobic.003G012800.1.p |



