 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sobic.003G096300.1.p | ||||||||
| Common Name | Sb03g008090, SORBIDRAFT_03g008090 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Andropogonodae; Andropogoneae; Sorghinae; Sorghum
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 816aa MW: 86894.2 Da PI: 5.153 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 70 | 2.9e-22 | 107 | 162 | 1 | 56 |
TT--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 1 rrkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+++ +++t++q++eLe+lF+++++p++++r eL+k+lgL+ rqVk+WFqNrR+++k
Sobic.003G096300.1.p 107 KKRYHRHTPQQIQELEALFKECPHPDEKQRGELSKRLGLDPRQVKFWFQNRRTQMK 162
678899***********************************************999 PP
| |||||||
| 2 | START | 182.3 | 2.7e-57 | 325 | 557 | 1 | 206 |
HHHHHHHHHHHHHHHHC-TT-EEEEEXCCTTEEEEEEESSS............SCEEEEEEEECCSCHHH.HHHHHHCCCGGCT-TT-S CS
START 1 elaeeaaqelvkkalaeepgWvkssesengdevlqkfeeskv...........dsgealrasgvvdmvla.llveellddkeqWdetla 77
ela +a++elvk+a+ +ep+W s +++++e l +e +++ + +ea+r+sg+v+ + lve+l+d + +W+ ++
Sobic.003G096300.1.p 325 ELAISAMDELVKLAQIDEPLWLPSLNGSPNKELLNFEEYAHSflpcvgvkpvgYVSEASRESGLVIFDNSlALVETLMDER-RWSDMFS 412
57889********************555555555555555555666679**************997655548999999999.******* PP
....EEEEEEEECTT......EEEEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE..-TTS--....-TTSEE-EESSE CS
START 78 ....kaetlevissg......galqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvd..seqkppe...sssvvRaellpS 150
ka++le ++sg g l lm+aelq+lsplvp R+++f+R+++ql +g w++vdvS+d ++++++ +++ +R+++lpS
Sobic.003G096300.1.p 413 cmiaKATVLEEVTSGiagsrnGGLLLMKAELQVLSPLVPiREVTFLRFCKQLAEGAWAVVDVSIDglVRDQNSAtasNAGNIRCRRLPS 501
****************************************************************954333443367899********** PP
EEEEEEECTCEEEEEEEE-EE--SSXXHHHHHHHHHHHHHHHHHHHHHHTXXXXXX CS
START 151 giliepksnghskvtwvehvdlkgrlphwllrslvksglaegaktwvatlqrqcek 206
g+++++++ng++kvtwveh++++++++h+l+r+l++sgla+ga++w+a lqrqce+
Sobic.003G096300.1.p 502 GCVMQDTPNGYCKVTWVEHTEYDEASVHQLYRPLLRSGLAFGARRWLAMLQRQCEC 557
******************************************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 1.2E-23 | 92 | 164 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 1.42E-21 | 95 | 164 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 18.285 | 104 | 164 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 2.8E-21 | 105 | 168 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 4.54E-20 | 106 | 164 | No hit | No description |
| Pfam | PF00046 | 9.7E-20 | 107 | 162 | IPR001356 | Homeobox domain |
| PROSITE pattern | PS00027 | 0 | 139 | 162 | IPR017970 | Homeobox, conserved site |
| PROSITE profile | PS50848 | 47.097 | 316 | 560 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 5.5E-31 | 318 | 557 | No hit | No description |
| CDD | cd08875 | 3.60E-113 | 320 | 556 | No hit | No description |
| SMART | SM00234 | 1.3E-45 | 325 | 557 | IPR002913 | START domain |
| Pfam | PF01852 | 4.8E-50 | 326 | 557 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 1.15E-21 | 577 | 784 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009827 | Biological Process | plant-type cell wall modification | ||||
| GO:0042335 | Biological Process | cuticle development | ||||
| GO:0043481 | Biological Process | anthocyanin accumulation in tissues in response to UV light | ||||
| GO:0048765 | Biological Process | root hair cell differentiation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 816 aa Download sequence Send to blast |
MSFGSLFDGG AGGGGGGGMQ FPFSTGFSSS PALSLGLDNA GGGGMVGRML PGGAGAGAAA 60 DGGGMMMGRD ADAENDSRSG SDHLDAMSAG GAEDEDDAEP GNPRKRKKRY HRHTPQQIQE 120 LEALFKECPH PDEKQRGELS KRLGLDPRQV KFWFQNRRTQ MKTQLERHEN ALLKQENDKL 180 RAENMAIREA MRSPMCGSCG SPAMLGEVSL EEQHLCIENA RLKDELSRVY ALATKFLGKP 240 MSILSAGTML QPNLSLPMPS SSLELAVGGG LRGLGSIPSA ATMPGSMGDF AGGVSSPLGT 300 VITPARTTGS APPPMVGIDR SMLLELAISA MDELVKLAQI DEPLWLPSLN GSPNKELLNF 360 EEYAHSFLPC VGVKPVGYVS EASRESGLVI FDNSLALVET LMDERRWSDM FSCMIAKATV 420 LEEVTSGIAG SRNGGLLLMK AELQVLSPLV PIREVTFLRF CKQLAEGAWA VVDVSIDGLV 480 RDQNSATASN AGNIRCRRLP SGCVMQDTPN GYCKVTWVEH TEYDEASVHQ LYRPLLRSGL 540 AFGARRWLAM LQRQCECLAI LMSPDTVSAN DSSVITQEGK RSMLKLARRM TENFCAGVSA 600 SSAREWSKLD GAAGSIGEDV RVMARKSVDE PGEPPGVVLS AATSVWVPVA PEKLFNFLRD 660 EQLRAEWDIL SNGGPMQEMA NIAKGQEHGN SVSLLRASAM SANQSSMLIL QETCTDASGS 720 MVVYAPVDIP AMQLVMNGGD STYVALLPSG FAILPDGPSG VGAEHKTGGS LLTVAFQILV 780 NSQPTAKLTV ESVETVNNLI SCTIKKIKTA LQCDA* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 102 | 108 | PRKRKKR |
| 2 | 103 | 107 | RKRKK |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Sbi.20121 | 0.0 | panicle | ||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00421 | DAP | Transfer from AT4G00730 | Download |
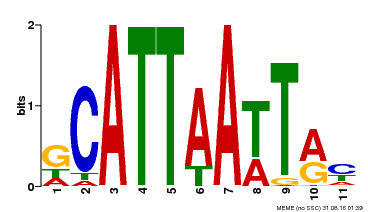
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Sobic.003G096300.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT063249 | 0.0 | BT063249.1 Zea mays full-length cDNA clone ZM_BFc0042P14 mRNA, complete cds. | |||
| GenBank | BT064453 | 0.0 | BT064453.1 Zea mays full-length cDNA clone ZM_BFc0170E20 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002457484.1 | 0.0 | homeobox-leucine zipper protein ROC5 | ||||
| Swissprot | Q6EPF0 | 0.0 | ROC5_ORYSJ; Homeobox-leucine zipper protein ROC5 | ||||
| TrEMBL | C5XEA6 | 0.0 | C5XEA6_SORBI; Uncharacterized protein | ||||
| STRING | Sb03g008090.1 | 0.0 | (Sorghum bicolor) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1203 | 37 | 116 | Representative plant | OGRP145 | 15 | 136 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G61150.1 | 0.0 | homeodomain GLABROUS 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Sobic.003G096300.1.p |
| Entrez Gene | 8079064 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



