 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sobic.004G027800.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Andropogonodae; Andropogoneae; Sorghinae; Sorghum
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 716aa MW: 80409 Da PI: 7.933 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 158 | 6.7e-49 | 106 | 246 | 4 | 134 |
DUF822 4 grkptwkErEnnkrRERrRRaiaakiyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyrkgskpl...eeaeaagssasas 89
g++++++E+E++k+RER+RRai+++++aGLR++Gn++lp+raD+n+Vl+AL+r+AGw+v++DGtt+r + +pl ++ g ++ as
Sobic.004G027800.1.p 106 GEREREREKERTKLRERHRRAITSRMLAGLRQHGNFPLPARADMNDVLAALARAAGWTVQPDGTTFRSSNQPLlppPPPQLHGVFQVAS 194
67899******************************************************************99988899********** PP
DUF822 90 pesslq.sslkssalaspvesysaspksssfpspssldsislasa......a 134
+e++ ++l+s+ + +p++s+++ +++++++spsslds+ + +
Sobic.004G027800.1.p 195 VETPALiNTLSSYVIGTPLDSQASALQTDDSLSPSSLDSVVADRSiktenyG 246
****9999*******************************9876555544443 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 1.5E-46 | 106 | 256 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| Gene3D | G3DSA:3.20.20.80 | 2.4E-171 | 275 | 709 | IPR013781 | Glycoside hydrolase, catalytic domain |
| SuperFamily | SSF51445 | 1.16E-167 | 276 | 711 | IPR017853 | Glycoside hydrolase superfamily |
| Pfam | PF01373 | 8.1E-83 | 298 | 670 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 9.5E-53 | 313 | 327 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 9.5E-53 | 334 | 352 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 9.5E-53 | 356 | 377 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 9.5E-53 | 449 | 471 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 9.5E-53 | 522 | 541 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 9.5E-53 | 556 | 572 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 9.5E-53 | 573 | 584 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 9.5E-53 | 591 | 614 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 9.5E-53 | 629 | 651 | IPR001554 | Glycoside hydrolase, family 14 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000272 | Biological Process | polysaccharide catabolic process | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0048831 | Biological Process | regulation of shoot system development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0016161 | Molecular Function | beta-amylase activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 716 aa Download sequence Send to blast |
MRVAPFDLLL TAVIERPRGK MDPLRSRSNC SWKEPTPRRS AAACGCGHAV ARRSRPQIGA 60 GAGGWKRPRA MKHPPQRDGE PSPSPPPQRR PRGFASTAGG SPRRRGERER EREKERTKLR 120 ERHRRAITSR MLAGLRQHGN FPLPARADMN DVLAALARAA GWTVQPDGTT FRSSNQPLLP 180 PPPPQLHGVF QVASVETPAL INTLSSYVIG TPLDSQASAL QTDDSLSPSS LDSVVADRSI 240 KTENYGNSSS VSSLNCMDND QLMRSAVLFP GDYTKTPYIP VYASLPMGII NSHCQLVDPE 300 SVRAELRHLK SLNVDGVVVD CWWGIVEAWT PRKYEWSGYR DLFGIIKEFK LKVQVVLSFH 360 GSGEIGSGDV LISLPKWIME IAKENQDIFF TDREGRRNTE CLSWGIDKER VLRGRTGIEV 420 YFDFMRSFHM EFRNLSEEGL VSSIEIGLGA SGELRYPSCP DTMGWKYPGI GEFQCYDRYM 480 QKHLRQSALS RGHLFWARGP DNAGYYNSRP HETGFFCDGG DYDSYYGRFF LNWYSGVLMD 540 HVDHVLSLAS LAFDGAEIVV KVPSIYWWYR TASHAAELTA GFYNPTNRDG YSPVFRMLKK 600 HSVILKLVCY GPEFTIQEND EAFADPEGLT WQVMNAAWDH GLSLCVESAL PCHNGEMYSQ 660 ILDTAKPRND PDRHHAAFFA YRQQPPFLLQ REVCFSELCT FVKCMHGEAP HNGEG* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wdq_A | 1e-121 | 278 | 708 | 11 | 444 | Beta-amylase |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00543 | DAP | Transfer from AT5G45300 | Download |
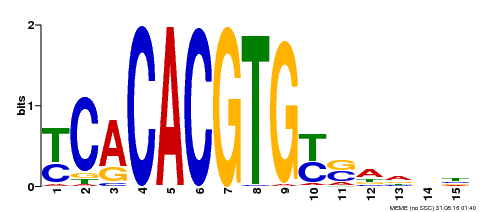
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Sobic.004G027800.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002451472.2 | 0.0 | beta-amylase 8 isoform X2 | ||||
| Swissprot | Q9FH80 | 0.0 | BAM8_ARATH; Beta-amylase 8 | ||||
| TrEMBL | A0A194YMC5 | 0.0 | A0A194YMC5_SORBI; Beta-amylase | ||||
| STRING | Sb04g002450.1 | 0.0 | (Sorghum bicolor) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G45300.2 | 0.0 | beta-amylase 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Sobic.004G027800.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



