 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sobic.004G251300.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Andropogonodae; Andropogoneae; Sorghinae; Sorghum
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 342aa MW: 37064.5 Da PI: 4.3829 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 64.5 | 1.5e-20 | 78 | 131 | 3 | 56 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
k++++t+eq++ Le+ Fe+++++ e+++eLA+klgL+ rqV vWFqNrRa++k
Sobic.004G251300.1.p 78 KKRRLTPEQVHLLERSFEEENKLEPERKTELARKLGLQPRQVAVWFQNRRARWK 131
5678*************************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 130.8 | 5.2e-42 | 77 | 168 | 1 | 92 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLre 89
ekkrrl+ eqv+lLE+sFeee+kLeperK+elar+LglqprqvavWFqnrRAR+ktkqlE+d+++Lk+++dal+++++ + +++++Lr+
Sobic.004G251300.1.p 77 EKKRRLTPEQVHLLERSFEEENKLEPERKTELARKLGLQPRQVAVWFQNRRARWKTKQLERDFDRLKASFDALRADHDVILQDNHRLRS 165
69**************************************************************************************9 PP
HD-ZIP_I/II 90 elk 92
++
Sobic.004G251300.1.p 166 QVV 168
986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 2.53E-20 | 71 | 135 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 18.22 | 73 | 133 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 2.0E-20 | 76 | 137 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 2.21E-20 | 78 | 134 | No hit | No description |
| Pfam | PF00046 | 7.8E-18 | 78 | 131 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 8.9E-22 | 79 | 140 | IPR009057 | Homeodomain-like |
| PRINTS | PR00031 | 2.2E-6 | 104 | 113 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 108 | 131 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 2.2E-6 | 113 | 129 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 6.3E-17 | 133 | 175 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0001558 | Biological Process | regulation of cell growth | ||||
| GO:0009637 | Biological Process | response to blue light | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009965 | Biological Process | leaf morphogenesis | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 342 aa Download sequence Send to blast |
MESGRLIFNA PGSGAGQMLF LDCGAAGGPG GGGGGGLFHR GGRPMLGLEE GRGVKRPFFT 60 SPDELLEEEY YDEQLPEKKR RLTPEQVHLL ERSFEEENKL EPERKTELAR KLGLQPRQVA 120 VWFQNRRARW KTKQLERDFD RLKASFDALR ADHDVILQDN HRLRSQVVSL TEKLQEKEAT 180 EGGASAATDA AAALPAVDDV KASLADDVEE PTEPAAEEEE AAFEVQQVKS EDRLSTGSGG 240 SAVVDTDALL YGAGCRFAAA VDSSVESYFP GGEDHHYHDC GMGPVNHGAG GGIQSDDDGA 300 GSDEGCSYYA EEEAAAFFAG HTHHHADDDE DAGQISWWMW N* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 125 | 133 | RRARWKTKQ |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Sbi.3331 | 1e-136 | callus| embryo| shoot | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in seedlings, stems, leaf sheaths and blades and panicles. {ECO:0000269|PubMed:17999151}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00323 | DAP | Transfer from AT3G01470 | Download |
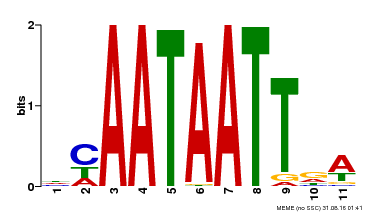
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Sobic.004G251300.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT067017 | 0.0 | BT067017.1 Zea mays full-length cDNA clone ZM_BFb0151L17 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021314788.1 | 0.0 | homeobox-leucine zipper protein HOX16-like isoform X1 | ||||
| Swissprot | A2X980 | 1e-143 | HOX16_ORYSI; Homeobox-leucine zipper protein HOX16 | ||||
| TrEMBL | A0A194YRG9 | 0.0 | A0A194YRG9_SORBI; Uncharacterized protein | ||||
| STRING | Sb04g029080.1 | 0.0 | (Sorghum bicolor) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2609 | 38 | 87 | Representative plant | OGRP129 | 16 | 189 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G01470.1 | 1e-61 | homeobox 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Sobic.004G251300.1.p |



