 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sobic.006G094500.1.p | ||||||||
| Common Name | Sb06g017920, SORBIDRAFT_06g017920 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Andropogonodae; Andropogoneae; Sorghinae; Sorghum
|
||||||||
| Family | Trihelix | ||||||||
| Protein Properties | Length: 409aa MW: 43139 Da PI: 6.7139 | ||||||||
| Description | Trihelix family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | trihelix | 41.5 | 3.6e-13 | 77 | 158 | 2 | 86 |
trihelix 2 WtkqevlaLiearremeerlrrgklkkplWeevskkm.rergfer...spkqCkekwenlnkrykkikegekkrtsessstcpyfdqle 86
W++ + aLi+a e+ +l+rg+l+ p+W+ev++++ ++ g+++ s +qCk++++ l+k+yk +++ ++ +++fd+l+
Sobic.006G094500.1.p 77 WSDGATSALIDAWGERFVALGRGSLRHPQWQEVADAVsSRDGYSKapkSDVQCKNRIDTLKKKYKVERAKPVSG-------WQFFDRLD 158
*************************************87788876222667**************999988875.......88888886 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF13837 | 2.0E-22 | 75 | 159 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009793 | Biological Process | embryo development ending in seed dormancy | ||||
| GO:0010431 | Biological Process | seed maturation | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 409 aa Download sequence Send to blast |
MDDDDDVSHS LSPSPPSSSA LPVDGPVTVA AAPPGSAMAV ALPIHRTAAS LYASATGGGG 60 GGGSGGGGGG GGREDAWSDG ATSALIDAWG ERFVALGRGS LRHPQWQEVA DAVSSRDGYS 120 KAPKSDVQCK NRIDTLKKKY KVERAKPVSG WQFFDRLDFL LAPTYGNKPG SGGNGGGGGH 180 NSNSRSQMPG ALRVGFPQRS RTPLMPAAGS AAKRRAPSPE PSVSSESSDG FPPVPALPAV 240 NGKRKRTDEG RADDGGSSGD DRAQGLRELA QAIRRFGEAY ERVEAAKLEQ AAEMERRRMD 300 FTQELESQRV QFFLNTQMEL TQAKNHASPA TAAVPAGGSS RRMSVVTDAG GSSNHHSRYR 360 ISHGDRHRHA PRPHYQQYHD NNNHAVAAAA SEGEQSDDEE DDYEEESQ* |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Sbi.4951 | 6e-83 | root | ||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00413 | DAP | Transfer from AT3G58630 | Download |
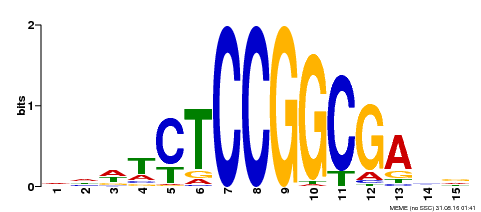
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Sobic.006G094500.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT068634 | 0.0 | BT068634.1 Zea mays full-length cDNA clone ZM_BFb0350B23 mRNA, complete cds. | |||
| GenBank | EU972739 | 0.0 | EU972739.1 Zea mays clone 387061 6b-interacting protein 1 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002447913.1 | 0.0 | trihelix transcription factor ASIL2 | ||||
| TrEMBL | C5Y986 | 0.0 | C5Y986_SORBI; Uncharacterized protein | ||||
| STRING | Sb06g017920.1 | 0.0 | (Sorghum bicolor) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2404 | 30 | 69 | Representative plant | OGRP487 | 16 | 84 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G14180.1 | 4e-33 | sequence-specific DNA binding transcription factors | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Sobic.006G094500.1.p |
| Entrez Gene | 8075868 |



