 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sobic.006G266600.1.p | ||||||||
| Common Name | Sb06g032880, SORBIDRAFT_06g032880 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Andropogonodae; Andropogoneae; Sorghinae; Sorghum
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 303aa MW: 31247.5 Da PI: 5.8737 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 28 | 5.1e-09 | 18 | 66 | 2 | 45 |
SSS-HHHHHHHHHHHHHTTTT......-HHHHHHHHTTTS-HHHHHHHHH CS
Myb_DNA-binding 2 grWTteEdellvdavkqlGgg......tWktIartmgkgRtlkqcksrwq 45
g+WT e ++ + +av+ gg +W++ a+ + g+t+++++ ++
Sobic.006G266600.1.p 18 GAWTREQEKAFENAVATTMGGeedgdaRWEKLAEAVE-GKTPEEVRRHYE 66
79**************999999***************.**********95 PP
| |||||||
| 2 | Myb_DNA-binding | 39.8 | 1.1e-12 | 129 | 173 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+WT++E+ l++ + +++G+g+W++I+r + Rt+ q+ s+ qky
Sobic.006G266600.1.p 129 AWTEDEHRLFLLGLEKYGKGDWRSISRNFVISRTPTQVASHAQKY 173
6*****************************99************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51293 | 7.495 | 15 | 73 | IPR017884 | SANT domain |
| SMART | SM00717 | 4.2E-5 | 16 | 71 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 4.7E-6 | 18 | 66 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 1.51E-8 | 20 | 75 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 9.04E-7 | 20 | 69 | No hit | No description |
| PROSITE profile | PS51294 | 16.605 | 122 | 178 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 2.09E-16 | 124 | 179 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 9.3E-11 | 126 | 176 | IPR001005 | SANT/Myb domain |
| TIGRFAMs | TIGR01557 | 2.8E-17 | 126 | 176 | IPR006447 | Myb domain, plants |
| CDD | cd00167 | 3.26E-10 | 129 | 174 | No hit | No description |
| Pfam | PF00249 | 3.4E-11 | 129 | 173 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 9.8E-11 | 129 | 172 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 303 aa Download sequence Send to blast |
MAVEEASSSG GGGEEGCGAW TREQEKAFEN AVATTMGGEE DGDARWEKLA EAVEGKTPEE 60 VRRHYELLVE DVDGIESGRV PLPTYAADGA AEEGGGGGGG GKKGGGGGGT HGDKGSAKSA 120 EQERRKGIAW TEDEHRLFLL GLEKYGKGDW RSISRNFVIS RTPTQVASHA QKYFIRLNSM 180 NRERRRSSIH DITSVNNGDA STAQGPITGQ TNGQAANPGK PSKQSPQPAN TPPGVDAYGT 240 TIGQPVGGPL VSAVGTPVTL PVSAPPHLAY GMHAPVPGAV VPGAPVNIAP MPYPMPPPSS 300 HG* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2cjj_A | 4e-17 | 20 | 89 | 11 | 77 | RADIALIS |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 99 | 107 | GGKKGGGGG |
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in young seedlings, developing leaves, sepals and trichomes. {ECO:0000269|PubMed:26243618}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that coordinates abscisic acid (ABA) biosynthesis and signaling-related genes via binding to the specific promoter motif 5'-(A/T)AACCAT-3'. Represses ABA-mediated salt (e.g. NaCl and KCl) stress tolerance. Regulates leaf shape and promotes vegetative growth. {ECO:0000269|PubMed:26243618}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00500 | DAP | Transfer from AT5G08520 | Download |
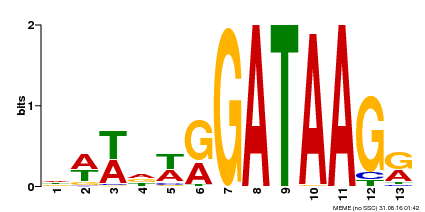
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Sobic.006G266600.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by salicylic acid (SA) and gibberellic acid (GA) (PubMed:16463103). Triggered by dehydration and salt stress (PubMed:26243618). {ECO:0000269|PubMed:16463103, ECO:0000269|PubMed:26243618}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | HF679443 | 0.0 | HF679443.1 Saccharum hybrid cultivar Co 86032 mRNA for ScMYB37 protein. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002448771.1 | 0.0 | transcription factor SRM1 | ||||
| Swissprot | Q9FNN6 | 1e-103 | SRM1_ARATH; Transcription factor SRM1 | ||||
| TrEMBL | C5YAI2 | 0.0 | C5YAI2_SORBI; Uncharacterized protein | ||||
| STRING | Sb06g032880.1 | 0.0 | (Sorghum bicolor) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP6376 | 38 | 54 | Representative plant | OGRP275 | 17 | 122 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G08520.1 | 1e-87 | MYB family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Sobic.006G266600.1.p |
| Entrez Gene | 8085866 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



