 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sobic.009G135000.1.p | ||||||||
| Common Name | Sb09g020110, SORBIDRAFT_09g020110 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Andropogonodae; Andropogoneae; Sorghinae; Sorghum
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 865aa MW: 94968.8 Da PI: 5.6897 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 111.3 | 5.9e-35 | 186 | 260 | 2 | 76 |
-SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S- CS
SBP 2 CqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkk 76
Cqv+gCead+ e+k yhrrh+vC +++a++v+++g+++r+CqqC++fh l +fDe+krsCrr+L++hn+rrr+k
Sobic.009G135000.1.p 186 CQVPGCEADIRELKGYHRRHRVCLRCAHAAAVMLDGVQKRYCQQCGKFHILLDFDEDKRSCRRKLERHNKRRRRK 260
************************************************************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 1.7E-26 | 181 | 247 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 29.487 | 183 | 260 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 3.27E-33 | 184 | 263 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 7.1E-27 | 186 | 260 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0055070 | Biological Process | copper ion homeostasis | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0016021 | Cellular Component | integral component of membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 865 aa Download sequence Send to blast |
MDVPDSGGGA PDAGEPIWDW GNLLDFVVQD DDSLVLPWDD AAAGIAAAAD PTEAATAPLL 60 PASPPLPQPV EVEPEPEPEP EPEPGPVLPP PPLRVQGIGR RVRKRDPRLV CPNYLAGRVP 120 CACPEVDEMV AAAEVEDVAT EFLAGARKKT KTAAAAARRS KAAAAGAAPG AAGGGAARAA 180 AAEMKCQVPG CEADIRELKG YHRRHRVCLR CAHAAAVMLD GVQKRYCQQC GKFHILLDFD 240 EDKRSCRRKL ERHNKRRRRK PDSKGTLDKE IDEQLDLSAD VSGDGELREE NMEGTTSEML 300 ETVLSNKVLD RETPVGSEDV LSSPTCTQPS LQNDQSKSVV TFAASVEACI GAKQESVKLV 360 NSPMHDTKSA YSSSCPTGRI SFKLYDWNPA EFPRRLRHQI FEWLASMPVE LEGYIRPGCT 420 ILTVFIAMPQ HMWDKLSDDA ADLLRNLVNS PNSLLLGKGA FFIHVNNMLF QVLKDGATLM 480 STRLDVQAPR IDYVHPTWFE AGKPGDLILY GSSLDQPNFR SLLSFDGDYL KHDCYRLTSH 540 DTFDRVENGD LIPDSQHEIF RINITQSRPD IHGPAFVEVE NIFGLSNFVP ILFGSKQLCS 600 ELERIQDALC GSYSKNNVLG ELLSASSDPH ERRKLHSSVM SGFLIDIGWL IRKTTPDEFK 660 NVLSSTNIQR WIHILKFLIQ NDFINVLEII VKSMDTIIGS EILSNLERGR LEDHVTTFLG 720 YVSHARNIVE RRANHDEKTQ IETGGIIVNS PNQPSLGASL PHASENTDIG GDNKLNSADE 780 EETMPLVTRD VSHRHCCQPD MTARWLKPSL IVTYPGGATR MRLVTTVVVA AVLCFTACLV 840 LFHPHGVGVL AAPVKRYLSS DSAS* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul5_A | 6e-41 | 185 | 265 | 5 | 85 | squamosa promoter binding protein-like 7 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 242 | 260 | KRSCRRKLERHNKRRRRKP |
| 2 | 246 | 258 | RRKLERHNKRRRR |
| 3 | 253 | 260 | NKRRRRKP |
| 4 | 254 | 259 | KRRRRK |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Sbi.12407 | 0.0 | ovary | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Ubiquitous. {ECO:0000269|PubMed:16861571}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00604 | PBM | Transfer from AT5G18830 | Download |
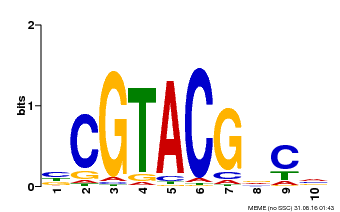
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Sobic.009G135000.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002439790.1 | 0.0 | squamosa promoter-binding-like protein 9 isoform X1 | ||||
| Swissprot | Q6I576 | 0.0 | SPL9_ORYSJ; Squamosa promoter-binding-like protein 9 | ||||
| TrEMBL | C5YXS3 | 0.0 | C5YXS3_SORBI; Uncharacterized protein | ||||
| STRING | Sb09g020110.1 | 0.0 | (Sorghum bicolor) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP6922 | 33 | 44 | Representative plant | OGRP4828 | 13 | 20 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G18830.2 | 1e-130 | squamosa promoter binding protein-like 7 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Sobic.009G135000.1.p |
| Entrez Gene | 8085123 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



