 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sobic.009G221400.1.p | ||||||||
| Common Name | Sb09g027530, SORBIDRAFT_09g027530 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Andropogonodae; Andropogoneae; Sorghinae; Sorghum
|
||||||||
| Family | RAV | ||||||||
| Protein Properties | Length: 409aa MW: 43036.6 Da PI: 10.0687 | ||||||||
| Description | RAV family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 39.4 | 1.5e-12 | 81 | 129 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkr.krfslgkfgtaeeAakaaiaarkkleg 55
s+ykGV + +grW A+I++ r +r++lg+f + eAa+a++ a+++++g
Sobic.009G221400.1.p 81 SKYKGVVPQP-NGRWGAQIYE------RhQRVWLGTFTGEAEAARAYDVAAQRFRG 129
89****9888.8*********......44**********99*************98 PP
| |||||||
| 2 | B3 | 94.9 | 5.6e-30 | 215 | 315 | 1 | 89 |
EEEE-..-HHHHTT-EE--HHH.HTT.............---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--T CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh.............ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLke 76
f+k+ tpsdv+k++rlv+pk++ae+h gg+ +++ l +ed +g++W+++++y+++s++yvltkGW++Fvk++gL +
Sobic.009G221400.1.p 215 FDKTVTPSDVGKLNRLVIPKQHAEKHfplqlpaaaaavvGGE-CKGVLLNFEDATGKVWRFRYSYWNSSQSYVLTKGWSRFVKEKGLHA 302
89***************************9999988886444.5899****************************************** PP
T-EEEEEE-SSSE CS
B3 77 gDfvvFkldgrse 89
gD v F+++ +
Sobic.009G221400.1.p 303 GDAVGFYRSAGGK 315
*******764433 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00018 | 7.18E-23 | 81 | 137 | No hit | No description |
| SuperFamily | SSF54171 | 1.77E-16 | 81 | 137 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 2.8E-8 | 81 | 129 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 3.2E-26 | 82 | 143 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 19.862 | 82 | 137 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 1.5E-19 | 82 | 137 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:2.40.330.10 | 7.3E-40 | 208 | 331 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 3.4E-30 | 213 | 323 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 1.48E-27 | 213 | 311 | No hit | No description |
| Pfam | PF02362 | 1.8E-26 | 215 | 316 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 1.2E-19 | 215 | 325 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 12.929 | 215 | 324 | IPR003340 | B3 DNA binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 409 aa Download sequence Send to blast |
MDSTSCLLDD ASSGASTGNK NPAPAPAAAT GGKPLQRVGS GASAVMDAAE PGAEADSGSG 60 GAGRATGGCG VVSGNGKLPS SKYKGVVPQP NGRWGAQIYE RHQRVWLGTF TGEAEAARAY 120 DVAAQRFRGR DAVTNFRPLA ESDPEAAVEL RFLASRTKAE VVDMLRKHTY GEELAQNRRA 180 FAAASPAASP PPAKNNNPAA SSSSPTAVTA REHLFDKTVT PSDVGKLNRL VIPKQHAEKH 240 FPLQLPAAAA AVVGGECKGV LLNFEDATGK VWRFRYSYWN SSQSYVLTKG WSRFVKEKGL 300 HAGDAVGFYR SAGGKQQFFI DCKLRPKTTT TAASFVNATT TTAAPSPVKA VRLFGVDLLT 360 TPRPGPAVVA APEQEEIAMA NKRARDAIAA CTPVHMVFKK QCIYFALT* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wid_A | 2e-50 | 212 | 333 | 11 | 125 | DNA-binding protein RAV1 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Sbi.19461 | 1e-129 | leaf| root | ||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00025 | PBM | Transfer from AT1G68840 | Download |
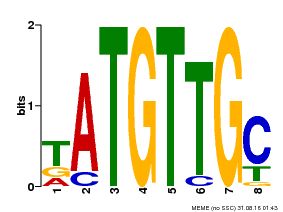
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Sobic.009G221400.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT041962 | 0.0 | BT041962.2 Zea mays full-length cDNA clone ZM_BFb0089O18 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021302722.1 | 0.0 | AP2/ERF and B3 domain-containing protein Os05g0549800 | ||||
| Swissprot | Q6L4H4 | 1e-179 | Y5498_ORYSJ; AP2/ERF and B3 domain-containing protein Os05g0549800 | ||||
| TrEMBL | A0A1B6P9U1 | 0.0 | A0A1B6P9U1_SORBI; Uncharacterized protein | ||||
| STRING | Sb09g027530.1 | 0.0 | (Sorghum bicolor) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP456 | 38 | 203 | Representative plant | OGRP365 | 16 | 108 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G25560.1 | 1e-100 | RAV family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Sobic.009G221400.1.p |
| Entrez Gene | 8061113 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



