 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sobic.009G231800.1.p | ||||||||
| Common Name | Sb09g028450, SORBIDRAFT_09g028450 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Andropogonodae; Andropogoneae; Sorghinae; Sorghum
|
||||||||
| Family | ARF | ||||||||
| Protein Properties | Length: 740aa MW: 80854.1 Da PI: 7.6664 | ||||||||
| Description | ARF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 77.9 | 1.1e-24 | 171 | 272 | 1 | 99 |
EEEE-..-HHHHTT-EE--HHH.HTT......---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEE CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh......ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFk 83
f+k+lt sd++++g +++p++ ae++ + +++ s++l+ +d++g +W++++iyr++++r++lt+GW+ Fv+ ++L +gD v+F
Sobic.009G231800.1.p 171 FCKTLTASDTSTHGGFSVPRRAAEDCfppldyS-QQRPSQELVAKDLHGTEWRFRHIYRGQPRRHLLTTGWSAFVNKKKLVSGDAVLFL 258
99****************************963.3445679************************************************ PP
E-SSSEE..EEEEE-S CS
B3 84 ldgrsefelvvkvfrk 99
+ +++el+++v+r+
Sobic.009G231800.1.p 259 --RGDDGELRLGVRRA 272
..3489999***9996 PP
| |||||||
| 2 | Auxin_resp | 111.3 | 9.5e-37 | 297 | 377 | 1 | 82 |
Auxin_resp 1 aahaastksvFevvYnPrastseFvvkvekvekalkvkvsvGmRfkmafetedsserrlsGtvvgvsdldpvrWpnSkWrsL 82
+aha++tksvF+++YnPr s+seF+++++k++k++++++s+G Rfkm++e++d+serr++G+++g+ d+dp W++SkW++L
Sobic.009G231800.1.p 297 VAHAVATKSVFHIYYNPRLSQSEFIIPYSKFMKSFSQQFSAGLRFKMRYESDDASERRCTGVIAGIGDADP-MWRGSKWKCL 377
79*********************************************************************.9********9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.40.330.10 | 6.0E-41 | 163 | 274 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 2.09E-43 | 164 | 286 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 5.76E-23 | 169 | 271 | No hit | No description |
| Pfam | PF02362 | 1.4E-22 | 171 | 272 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 5.3E-23 | 171 | 273 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 13.493 | 171 | 273 | IPR003340 | B3 DNA binding domain |
| Pfam | PF06507 | 2.5E-31 | 297 | 377 | IPR010525 | Auxin response factor |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009734 | Biological Process | auxin-activated signaling pathway | ||||
| GO:0010050 | Biological Process | vegetative phase change | ||||
| GO:0010158 | Biological Process | abaxial cell fate specification | ||||
| GO:0010582 | Biological Process | floral meristem determinacy | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 740 aa Download sequence Send to blast |
MAGIDLNDTV EEDEEEAEPG NPACSQQSRT SSAATPPPPP LTPLLQPRPG AAVCLELWHA 60 CAGPVAPLPR KGTVVVYLPQ GHLEHLGDAA AAAAGGAPAP AALPPHVFCR VVDVTLHADA 120 STDEVYAQLA LVAENEDVAR RLRGGSEDGS AGDGDDGEAV KQRFSRMPHM FCKTLTASDT 180 STHGGFSVPR RAAEDCFPPL DYSQQRPSQE LVAKDLHGTE WRFRHIYRGQ PRRHLLTTGW 240 SAFVNKKKLV SGDAVLFLRG DDGELRLGVR RAAQLKNGSA FPALYNQCSN LGSLANVAHA 300 VATKSVFHIY YNPRLSQSEF IIPYSKFMKS FSQQFSAGLR FKMRYESDDA SERRCTGVIA 360 GIGDADPMWR GSKWKCLMVR WDDDVDFRRP NRISPWEIEL TSSVSGSHLS APNAKRLKPC 420 LPPDYLVPNG SGCPDFAESA QFHKVLQGQE LLGYRTRDNA AVATSQPCEA RNMQYIDERS 480 CSNNVSNSIP GVPRIGVRTP LGNPRFSYHC SGFGESPRFQ KVLQGQEVFQ PYRGTLVDPS 540 LRNSGFHQQD GSHVPTQANK WHPQLHGCAF RGTQAPAIPS QSSSPPSVLM FQRDNPKMSP 600 FEFGHCHMDK NEDMRAMFGH AGGIGRTEQT MMLQAHNVSG GMGNRDVTVE KFQPTVAVGR 660 DGSDNREVTK NSCKIFGISL TEKVPAIKEK DCGDTNYPSP FLSLKQHVPK SLGNSCATIH 720 EQRPVVGRVI DVSTVDMMI* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ldv_A | 1e-113 | 52 | 407 | 18 | 361 | Auxin response factor 1 |
| 4ldw_A | 1e-113 | 52 | 407 | 18 | 361 | Auxin response factor 1 |
| 4ldw_B | 1e-113 | 52 | 407 | 18 | 361 | Auxin response factor 1 |
| 4ldx_A | 1e-113 | 52 | 407 | 18 | 361 | Auxin response factor 1 |
| 4ldx_B | 1e-113 | 52 | 407 | 18 | 361 | Auxin response factor 1 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Sbi.12623 | 0.0 | embryo| panicle | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in roots, culms, leaves and young panicles. {ECO:0000269|PubMed:17408882}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Auxin response factors (ARFs) are transcriptional factors that bind specifically to the DNA sequence 5'-TGTCTC-3' found in the auxin-responsive promoter elements (AuxREs). | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00021 | PBM | Transfer from AT2G33860 | Download |
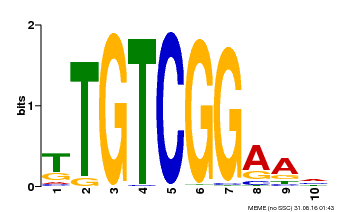
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Sobic.009G231800.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002440246.1 | 0.0 | auxin response factor 15 | ||||
| Swissprot | Q8S985 | 0.0 | ARFO_ORYSJ; Auxin response factor 15 | ||||
| TrEMBL | C5YVJ4 | 0.0 | C5YVJ4_SORBI; Auxin response factor | ||||
| STRING | Sb09g028450.1 | 0.0 | (Sorghum bicolor) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2537 | 35 | 80 | Representative plant | OGRP6685 | 10 | 18 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G33860.1 | 1e-159 | ARF family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Sobic.009G231800.1.p |
| Entrez Gene | 8074594 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



