 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sobic.010G096300.1.p | ||||||||
| Common Name | Sb10g008400, SORBIDRAFT_10g008400 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Andropogonodae; Andropogoneae; Sorghinae; Sorghum
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 467aa MW: 50071.5 Da PI: 4.8283 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 87.4 | 1.4e-27 | 189 | 243 | 1 | 56 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRla 56
k++++Wtp+LH+rFv+aveqL G++kA+P++ile+m++++Lt+++++SHLQkYR++
Sobic.010G096300.1.p 189 KAKVDWTPDLHRRFVQAVEQL-GIDKAVPSRILEIMGIDSLTRHNIASHLQKYRSH 243
6799*****************.********************************86 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 16.334 | 186 | 245 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 4.3E-18 | 187 | 246 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 3.0E-27 | 187 | 246 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.6E-26 | 189 | 243 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 2.2E-7 | 192 | 241 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0007165 | Biological Process | signal transduction | ||||
| GO:0009658 | Biological Process | chloroplast organization | ||||
| GO:0009910 | Biological Process | negative regulation of flower development | ||||
| GO:0010380 | Biological Process | regulation of chlorophyll biosynthetic process | ||||
| GO:0010638 | Biological Process | positive regulation of organelle organization | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:1900056 | Biological Process | negative regulation of leaf senescence | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 467 aa Download sequence Send to blast |
MCADVEECGG AAAGIKEMEE IAVGPVSDLD FDFTVDDIDF GDFFLRLDDD DRDALPDLEV 60 DPAEIFITDF EAIATGAGHG GVMDQEVPSV LPLADAVHIG AVDVDVDPCC PGVLGGEEDN 120 VTTCEDVEEG KGECNDDHAE GEEVVAGNGD SGEGGCGTVL GEKSPSSTTS QEAESRHKSS 180 NKHSHGKKKA KVDWTPDLHR RFVQAVEQLG IDKAVPSRIL EIMGIDSLTR HNIASHLQKY 240 RSHRKHMLAR EVEAATWTHR RQMYAAGGAP NGVKRPDSNA WTVPTIGFPP PPPPPPPPPP 300 PSPHPMQHFG RPLHVWGHPT PTVESPRVPM WPRHLVPRTP PPPWAPPPPS DPAFWHHAYM 360 RGPAHMPGQV TPCVAVPMPA ARFPAPPVRG ALPCPPPMYR PLVPPTLDTQ FQLQTQPSSE 420 SIDAAIGDVL TKPWLPLPLG LKPPSVDSVM GELQRQGVAN VPPACG* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1irz_A | 2e-16 | 186 | 241 | 2 | 57 | ARR10-B |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Sbi.8118 | 0.0 | leaf| shoot | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in leaves. {ECO:0000269|PubMed:11340194}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcriptional activator that promotes chloroplast development. Acts as an activator of nuclear photosynthetic genes involved in chlorophyll biosynthesis, light harvesting, and electron transport. {ECO:0000269|PubMed:19808806}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00022 | PBM | Transfer from AT2G20570 | Download |
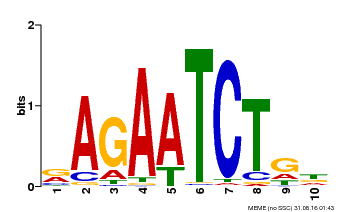
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Sobic.010G096300.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By light. {ECO:0000269|PubMed:11340194}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AF318580 | 1e-100 | AF318580.1 Zea mays putative transcription factor ZmGLK1 (Glk1) mRNA, complete cds. | |||
| GenBank | BT055220 | 1e-100 | BT055220.1 Zea mays full-length cDNA clone ZM_BFc0139J21 mRNA, complete cds. | |||
| GenBank | KJ726818 | 1e-100 | KJ726818.1 Zea mays clone pUT3357 G2-like transcription factor (GLK57) mRNA, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002436765.1 | 0.0 | probable transcription factor GLK1 | ||||
| Swissprot | Q5Z5I4 | 1e-150 | GLK1_ORYSJ; Probable transcription factor GLK1 | ||||
| TrEMBL | C5Z7F7 | 0.0 | C5Z7F7_SORBI; Uncharacterized protein | ||||
| STRING | Sb10g008400.1 | 0.0 | (Sorghum bicolor) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2415 | 38 | 86 | Representative plant | OGRP2840 | 15 | 29 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G20570.1 | 2e-59 | GBF's pro-rich region-interacting factor 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Sobic.010G096300.1.p |
| Entrez Gene | 8057890 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



