 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Solyc06g064540.1.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Solaneae; Solanum; Lycopersicon
|
||||||||
| Family | LBD | ||||||||
| Protein Properties | Length: 149aa MW: 16955.5 Da PI: 8.5937 | ||||||||
| Description | LBD family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF260 | 124.6 | 5e-39 | 13 | 113 | 1 | 101 |
DUF260 1 aCaaCkvlrrkCakdCvlapyfpaeqpkkfanvhklFGasnvlkllkalpeeeredamsslvyeAearardPvyGavgvilklqqqleqlk 91
+CaaCk lrr+C++dC++ pyfp ++p++f +vh+++Gasnv kll++++e++r d+++sl+yeA +r++ PvyG+vg+i+ l+q++++l+
Solyc06g064540.1.1 13 RCAACKQLRRRCPSDCIFLPYFPPNNPQRFSFVHRIYGASNVGKLLQQVQEHQRADVADSLYYEAYCRIKSPVYGCVGIITILHQEIDRLE 103
6****************************************************************************************** PP
DUF260 92 aelallkeel 101
el+++++e+
Solyc06g064540.1.1 104 RELVKVQAEI 113
*****99875 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50891 | 24.406 | 12 | 113 | IPR004883 | Lateral organ boundaries, LOB |
| Pfam | PF03195 | 1.8E-38 | 13 | 110 | IPR004883 | Lateral organ boundaries, LOB |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0016020 | Cellular Component | membrane | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 149 aa Download sequence Send to blast |
MVDLQNVLMN SRRCAACKQL RRRCPSDCIF LPYFPPNNPQ RFSFVHRIYG ASNVGKLLQQ 60 VQEHQRADVA DSLYYEAYCR IKSPVYGCVG IITILHQEID RLERELVKVQ AEISLVKGQT 120 QIEGQLAQGQ QVQLSSSSFH LPTTTPWF* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5ly0_A | 4e-37 | 14 | 113 | 12 | 111 | LOB family transfactor Ramosa2.1 |
| 5ly0_B | 4e-37 | 14 | 113 | 12 | 111 | LOB family transfactor Ramosa2.1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00381 | DAP | Transfer from AT3G26620 | Download |
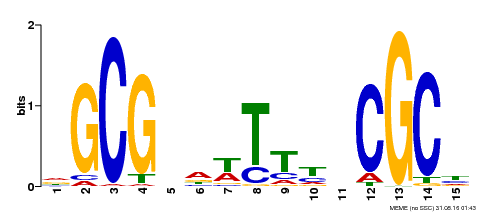
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Solyc06g064540.1.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_027773287.1 | 7e-99 | LOB domain-containing protein 24-like | ||||
| Swissprot | P59467 | 7e-47 | LBD23_ARATH; LOB domain-containing protein 23 | ||||
| Swissprot | P59468 | 5e-47 | LBD24_ARATH; LOB domain-containing protein 24 | ||||
| TrEMBL | A0A3Q7HRS9 | 1e-100 | A0A3Q7HRS9_SOLLC; Uncharacterized protein | ||||
| STRING | Solyc06g064540.1.1 | 1e-107 | (Solanum lycopersicum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA5277 | 20 | 39 | Representative plant | OGRP60 | 16 | 318 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G26660.1 | 2e-49 | LOB domain-containing protein 24 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Solyc06g064540.1.1 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



