 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Solyc06g075510.2.1 | ||||||||
| Common Name | LOC100736535 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Solaneae; Solanum; Lycopersicon
|
||||||||
| Family | AP2 | ||||||||
| Protein Properties | Length: 475aa MW: 52534.1 Da PI: 7.9137 | ||||||||
| Description | AP2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 53.2 | 7.2e-17 | 151 | 200 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkr.krfslgkfgtaeeAakaaiaarkkleg 55
s+y+GV++++++grW+++I+d + k+++lg f+ta Aa+a+++a+ k++g
Solyc06g075510.2.1 151 SQYRGVTFYRRTGRWESHIWD------CgKQVYLGGFDTAHTAARAYDRAAIKFRG 200
78*******************......55************************997 PP
| |||||||
| 2 | AP2 | 39.3 | 1.6e-12 | 243 | 291 | 1 | 53 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkl 53
s+++GV+ +k grW+A+ + +k+++lg f+++ eAa+a+++a+ k
Solyc06g075510.2.1 243 SKFRGVTLHK-CGRWEARMGQF--L-GKKYIYLGLFDSEVEAARAYDKAAIKT 291
79********.7******5553..2.26**********99**********886 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF00847 | 7.1E-9 | 151 | 200 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 9.15E-16 | 151 | 209 | IPR016177 | DNA-binding domain |
| SMART | SM00380 | 3.5E-32 | 152 | 214 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 17.662 | 152 | 208 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 4.1E-17 | 152 | 209 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 1.42E-23 | 152 | 207 | No hit | No description |
| Pfam | PF00847 | 1.2E-7 | 243 | 290 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 6.57E-24 | 243 | 303 | No hit | No description |
| SuperFamily | SSF54171 | 3.73E-16 | 243 | 302 | IPR016177 | DNA-binding domain |
| SMART | SM00380 | 6.2E-30 | 244 | 307 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 15.132 | 244 | 301 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 4.8E-15 | 244 | 301 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 2.6E-5 | 245 | 256 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 2.6E-5 | 283 | 303 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 475 aa Download sequence Send to blast |
MLDLNVSVIY NNDLPQVSLL DESATSNSSL RNAEATTSAG DEDSCAGELF AFNFGILKVE 60 GAETSRSSNN DDEEGYGKNQ RVTHSQFVTR QLFPVDDGEL NRKQTDRVIL SSARSGTSIG 120 FGDVRIIQQQ QTEQPKQQVK KSRRGPRSRS SQYRGVTFYR RTGRWESHIW DCGKQVYLGG 180 FDTAHTAARA YDRAAIKFRG VDADINFSLS DYEEDMQQMK NLGKEEFVHL LRRHSTGFSR 240 GSSKFRGVTL HKCGRWEARM GQFLGKKYIY LGLFDSEVEA ARAYDKAAIK TSGREAVTNF 300 EPSSYEGETM SLPQSEGSQH DLDLNLGIST TSSKENDRLG GSRYHPYDMQ DATKPKMDKP 360 GSVIVGSSHL KGLPMSSQQA QLWTGIYSNF SSSYEGRAYD KRKDTGSSQG PPNWALQMPS 420 QVDTNSPLTM FCTAASSGFF IPSTTSITSS TSALATSTNA SQCFYQINPR LPLP* |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Les.740 | 0.0 | leaf | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Detected in developing branch and spikelet meristems at very early stages of inflorescence formation, and is maintained uniformly in the spikelet meristems until floral organs initiation. Present in some floral organ primordia. {ECO:0000269|PubMed:22003982}. | |||||
| Uniprot | TISSUE SPECIFICITY: Highly expressed in developing panicles and in young seedlings (PubMed:20017947, PubMed:22003982). Present at low levels at all developmental stages (PubMed:22003982). {ECO:0000269|PubMed:20017947, ECO:0000269|PubMed:22003982}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor (By similarity). Involved in spikelet transition (Probable). Together with SNB, controls synergistically inflorescence architecture and floral meristem establishment via the regulation of spatio-temporal expression of B- and E-function floral organ identity genes in the lodicules and of spikelet meristem genes (PubMed:22003982). Prevents lemma and palea elongation as well as grain growth (PubMed:28066457). {ECO:0000250|UniProtKB:P47927, ECO:0000269|PubMed:22003982, ECO:0000269|PubMed:28066457, ECO:0000305|PubMed:26631749}. | |||||
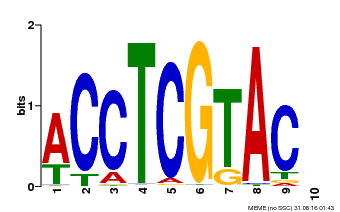
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00615 | PBM | Transfer from AT2G28550 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Solyc06g075510.2.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Target of miR172 microRNA mediated cleavage, particularly during floral organ development. {ECO:0000269|PubMed:22003982, ECO:0000305|PubMed:20017947, ECO:0000305|PubMed:26631749, ECO:0000305|PubMed:28066457}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | HQ586954 | 0.0 | Solanum lycopersicum AP2 transcription factor SlAP2e mRNA, complete cds | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_001233891.2 | 0.0 | APETALA2-like protein AP2e | ||||
| Swissprot | Q84TB5 | 1e-102 | AP22_ORYSJ; APETALA2-like protein 2 | ||||
| TrEMBL | F6KQN5 | 0.0 | F6KQN5_SOLLC; AP2 transcription factor SlAP2e | ||||
| STRING | Solyc06g075510.2.1 | 0.0 | (Solanum lycopersicum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA591 | 24 | 116 | Representative plant | OGRP497 | 17 | 84 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G28550.3 | 1e-108 | related to AP2.7 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Solyc06g075510.2.1 |
| Entrez Gene | 100736535 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



