 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Solyc12g007180.1.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Solaneae; Solanum; Lycopersicon
|
||||||||
| Family | CPP | ||||||||
| Protein Properties | Length: 499aa MW: 54518.7 Da PI: 7.3274 | ||||||||
| Description | CPP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | TCR | 45.9 | 1.1e-14 | 87 | 126 | 2 | 42 |
TCR 2 ekkgCnCkkskClkkYCeCfaagkkCseeCkCedCkNkeek 42
++k+CnCk+s+Clk+YCeCfa+g++C+ C+C++C+N+ e+
Solyc12g007180.1.1 87 KQKQCNCKHSRCLKLYCECFASGVYCDG-CNCNNCHNNVEN 126
89**************************.********9875 PP
| |||||||
| 2 | TCR | 51.2 | 2.5e-16 | 171 | 210 | 1 | 40 |
TCR 1 kekkgCnCkkskClkkYCeCfaagkkCseeCkCedCkNke 40
k++kgC+Ckks ClkkYCeCf+a+ Cse+CkC dCkN e
Solyc12g007180.1.1 171 KHNKGCHCKKSGCLKKYCECFQANILCSENCKCMDCKNFE 210
589***********************************65 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01114 | 7.1E-16 | 86 | 126 | IPR033467 | Tesmin/TSO1-like CXC domain |
| PROSITE profile | PS51634 | 37.088 | 87 | 211 | IPR005172 | CRC domain |
| Pfam | PF03638 | 1.6E-11 | 88 | 123 | IPR005172 | CRC domain |
| SMART | SM01114 | 8.4E-20 | 171 | 212 | IPR033467 | Tesmin/TSO1-like CXC domain |
| Pfam | PF03638 | 9.2E-13 | 174 | 209 | IPR005172 | CRC domain |
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 499 aa Download sequence Send to blast |
MSVFSCIPVE TGICEWPRGK KMFGLPFCSF SFDPVWLSVF IFWLICEAIH DFRDDNHDPL 60 LATLKPESPR ARPRQSASEV KDGTPKKQKQ CNCKHSRCLK LYCECFASGV YCDGCNCNNC 120 HNNVENEPAR REAVEATLER NPHAFRPKIA SSPHGARDNK EEAGDGLVLA KHNKGCHCKK 180 SGCLKKYCEC FQANILCSEN CKCMDCKNFE GSEERQALFH GDHANNMAYL QQAANAAITG 240 AIGSSGYGSP PVNKKRKSQE LFFGSTIKDP IHRHGQLQQG NHIKSSVLPS LSSSPGPCVG 300 NAATVGPSKF TYRSLLEDLI QPQDLKELCS VLVVYSGEAA KMLADEKDAS KKQAEGQTQA 360 SLASSTQEQV QNQKDSGSEK DMLDCSLSGD QGEKISMKES SLDGADVSKR RPMSPGTLAL 420 MCDESDSMFA AAITSPDDLA TLGCNTSSQL PHGQGMTETY AEQERIVLTK FRDCLNRLVT 480 LGEIKGRGKK MFFNGAGY* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5fd3_A | 4e-32 | 79 | 220 | 2 | 132 | Protein lin-54 homolog |
| 5fd3_B | 4e-32 | 79 | 220 | 2 | 132 | Protein lin-54 homolog |
| Search in ModeBase | ||||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Ubiquitous but expressed mostly in flowers. {ECO:0000269|PubMed:18057042}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Plays a role in development of both male and female reproductive tissues. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00045 | PBM | Transfer from AT4G29000 | Download |
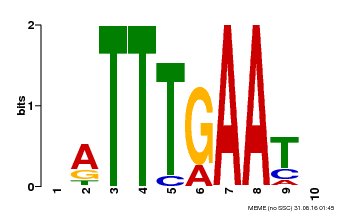
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Solyc12g007180.1.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004251614.1 | 0.0 | protein tesmin/TSO1-like CXC 5 isoform X2 | ||||
| Swissprot | Q9SZD1 | 1e-178 | TCX5_ARATH; Protein tesmin/TSO1-like CXC 5 | ||||
| TrEMBL | A0A3Q7J2R3 | 0.0 | A0A3Q7J2R3_SOLLC; Uncharacterized protein | ||||
| STRING | Solyc12g007180.1.1 | 0.0 | (Solanum lycopersicum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA2401 | 24 | 55 | Representative plant | OGRP1469 | 17 | 45 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G29000.1 | 1e-169 | Tesmin/TSO1-like CXC domain-containing protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Solyc12g007180.1.1 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



