 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sopen01g047740.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Solaneae; Solanum; Lycopersicon
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 1049aa MW: 118478 Da PI: 5.4465 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 178.1 | 1.1e-55 | 20 | 136 | 3 | 118 |
CG-1 3 ke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptf 94
+e ++rwl++ ei++iL n++k++lt e++ rp sgs++L++rk++ryfrkDG++w+kkkdgktv+E+hekLKvg+++vl+cyYah+ee+ +f
Sopen01g047740.1 20 SEvQHRWLRPAEICEILRNHRKFHLTPEAPFRPVSGSVFLFDRKVLRYFRKDGHNWRKKKDGKTVKEAHEKLKVGSIDVLHCYYAHGEEDDNF 112
5559***************************************************************************************** PP
CG-1 95 qrrcywlLeeelekivlvhylevk 118
qrr+yw+Le++l +iv+vhylevk
Sopen01g047740.1 113 QRRSYWMLEQDLMHIVFVHYLEVK 136
*********************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 80.494 | 15 | 141 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 1.1E-78 | 18 | 136 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 9.5E-49 | 22 | 135 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:2.60.40.10 | 2.2E-6 | 459 | 548 | IPR013783 | Immunoglobulin-like fold |
| SuperFamily | SSF81296 | 3.27E-17 | 463 | 549 | IPR014756 | Immunoglobulin E-set |
| Pfam | PF01833 | 2.6E-7 | 463 | 548 | IPR002909 | IPT domain |
| SuperFamily | SSF48403 | 3.11E-15 | 650 | 753 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 3.2E-14 | 657 | 755 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 1.04E-12 | 661 | 751 | No hit | No description |
| PROSITE profile | PS50297 | 16.6 | 663 | 748 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 0.003 | 696 | 725 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 9.778 | 696 | 728 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 2700 | 735 | 764 | IPR002110 | Ankyrin repeat |
| SMART | SM00015 | 5.8 | 870 | 892 | IPR000048 | IQ motif, EF-hand binding site |
| SuperFamily | SSF52540 | 8.23E-7 | 870 | 921 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| PROSITE profile | PS50096 | 8.004 | 871 | 900 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.003 | 872 | 891 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0072 | 893 | 915 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.157 | 894 | 918 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 7.8E-4 | 896 | 915 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0071275 | Biological Process | cellular response to aluminum ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1049 aa Download sequence Send to blast |
MEDCGSDPPG FRLDITQILS EVQHRWLRPA EICEILRNHR KFHLTPEAPF RPVSGSVFLF 60 DRKVLRYFRK DGHNWRKKKD GKTVKEAHEK LKVGSIDVLH CYYAHGEEDD NFQRRSYWML 120 EQDLMHIVFV HYLEVKGNKV NVSSIRSTKS VHPNYLNDCS LSDSFPTRHK KLTSANADST 180 SLASTLTEAH EEAESEDSHQ ACSRFHSYPD RASGMDSHLV ENRDTISSSY GSPQSSVEYT 240 PLPGIDGSGK CDLGNFASGP QRTIDLGSWE PLPQHCSNGE MVCQDDFKNN LSVHGNWQYS 300 FGQSPLQFHG QNVNQDLIAD SSYDLGLPWD LLTVRGPSYL CSNEKEEQLT QLNLQFLKSL 360 VEVQGDINQE NSMDMLELGD YSTIKQPHLS SVKVEEGLKK VDSFSRWVAK ELEDVEELHM 420 QPSNQMSWNV IDTEEEGSCL PSQLHVDSDS LNLSLSQEQV FSIIDFSPNW AYSNLETKIL 480 ITGRFLKSEG ELVEYKWSCM FGEVEVPAEV LADGVLRCHA PPHKPGVLPF YVTCSNRLAC 540 SEVREFEYRF GPYQEVGAAD VSMTEKHLLE RIENLLSLGP VSSCRSSDSM EDSEEKRSTV 600 NKIISMMEEE NKQIIERASY GDTSQCRVKE DLYFERKLKQ NFYAWLVHQV TDDGRGRTLL 660 DGEGQGVLHL VAALGYDWAF KPILASGLSV DFRDMNGWTA LHWAAFYGRE KTVVSLVSLG 720 ASPGALTDPS AEFPLGRTPA DLASANGHKG ISGFVAESSL TTHLSKLTVT DAKEELDSEV 780 CEAKVGETVT ERVAVSTTES DVPDVLSLKD SLAAIRNATQ AAARIHQIFR VQSFQRKQII 840 ENCDNELSSD ENAIAIVASR ACKLGQNNGI AHAAAIQIQK KFRGWNKRKE FLLIRQKIVQ 900 IQAHIRGHQV RKKYKPIIWS VGILEKVILR WRRKRSGLRG FRSEAVMSKP STQEDSLPED 960 DYDFLKEGRK QTEVRMQKAL ARVKSMTQYS EGRAQYRRLL TAAEGLREVK QDGPIQIPEI 1020 PEDTIYPEEE LFDVDSLLDD DTFMSIAFE |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in freezing tolerance in association with CAMTA1 and CAMTA3. Contributes together with CAMTA1 and CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:23581962). Involved together with CAMTA3 and CAMTA4 in the positive regulation of a general stress response (PubMed:25039701). Involved in tolerance to aluminum. Binds to the promoter of ALMT1 transporter and contributes to the positive regulation of aluminum-induced expression of ALMT1 (PubMed:25627216). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:23581962, ECO:0000269|PubMed:25039701, ECO:0000269|PubMed:25627216, ECO:0000305|PubMed:11925432}. | |||||
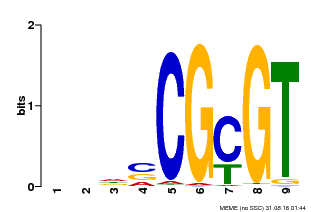
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00043 | PBM | Transfer from AT5G64220 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By salt, wounding, abscisic acid, H(2)O(2) and salicylic acid (PubMed:12218065). Induced by aluminum (PubMed:25627216). {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:25627216}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | JN558810 | 0.0 | JN558810.1 Solanum lycopersicum calmodulin-binding transcription factor SR1L mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015062177.1 | 0.0 | calmodulin-binding transcription activator 2 isoform X1 | ||||
| Swissprot | Q6NPP4 | 0.0 | CMTA2_ARATH; Calmodulin-binding transcription activator 2 | ||||
| TrEMBL | A0A3Q7ER35 | 0.0 | A0A3Q7ER35_SOLLC; Uncharacterized protein | ||||
| STRING | Solyc01g105230.2.1 | 0.0 | (Solanum lycopersicum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA2230 | 21 | 46 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G64220.2 | 0.0 | Calmodulin-binding transcription activator protein with CG-1 and Ankyrin domains | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



