 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sopen06g016880.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Solaneae; Solanum; Lycopersicon
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 378aa MW: 42927.6 Da PI: 10.067 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 102.1 | 3.4e-32 | 70 | 123 | 2 | 55 |
G2-like 2 prlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
prlrWtpeLH rFv+ave+LGG+++AtPk +l++m++kgL+++hvkSHLQ+YR+
Sopen06g016880.1 70 PRLRWTPELHLRFVHAVERLGGQDRATPKLVLQFMNIKGLSIAHVKSHLQMYRS 123
8****************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 12.917 | 66 | 126 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 5.5E-30 | 66 | 124 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 5.38E-16 | 67 | 125 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 6.7E-23 | 70 | 125 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 6.7E-10 | 71 | 122 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 378 aa Download sequence Send to blast |
MKGGSSSTEE DSKSNPKQQI EFEEEEDLET EVNSKAQGTT RSSSSNSTVE ENGKKPNSGS 60 VRQYVRSKTP RLRWTPELHL RFVHAVERLG GQDRATPKLV LQFMNIKGLS IAHVKSHLQM 120 YRSKKTDDPN QISTDGRFLL ENGDHHIFNL TQLSRLQGFN QTSSSSLRYD NALWNRQANS 180 LYNPYNMNIG GGSSISTRHG FASHVNGHSS HEQLFPWNKS IEIEKNQNRH NQRFWPPITQ 240 IGTSSAKQNL LMTLPMGNRN RDKDEIVKQF NRRSSEQEIM TQIRAKENNS LSSRGKWWMS 300 EEVVGTTTIN TSSSNKRKIQ DSDIDLDLNL SLKTTREYHQ KRLKGEQVGD LLSLSLFSSS 360 TTSEENNAKK ASTLDLTL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4r_A | 9e-17 | 71 | 125 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 9e-17 | 71 | 125 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 9e-17 | 71 | 125 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 9e-17 | 71 | 125 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00301 | DAP | Transfer from AT2G40260 | Download |
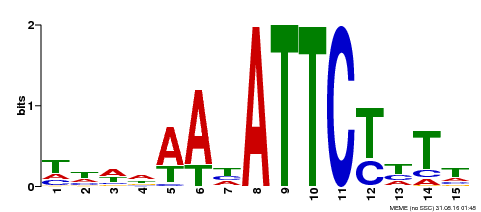
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | HG975445 | 0.0 | HG975445.1 Solanum pennellii chromosome ch06, complete genome. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015077982.1 | 0.0 | putative two-component response regulator ARR13 isoform X2 | ||||
| TrEMBL | A0A3Q7HNH9 | 0.0 | A0A3Q7HNH9_SOLLC; Uncharacterized protein | ||||
| STRING | PGSC0003DMT400077217 | 0.0 | (Solanum tuberosum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA8573 | 21 | 29 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G38300.1 | 6e-48 | G2-like family protein | ||||



