 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sopen08g001780.2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Solaneae; Solanum; Lycopersicon
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 667aa MW: 75452.6 Da PI: 6.1768 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 159.3 | 2.6e-49 | 60 | 202 | 3 | 143 |
DUF822 3 sgrkptwkErEnnkrRERrRRaiaakiyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyrkgskpleeaeaagssasaspesslq 95
+ rk+++kE+E++k+RER+RRai+++++aGLR++Gn++lp+raD+n+Vl+AL+r+AGw+ve+DGtt+r+ p ++a+ +g+ + +s+es+++
Sopen08g001780.2 60 KSRKEREKEKERTKLRERHRRAITSRMLAGLRQYGNFPLPVRADMNDVLAALARQAGWTVEPDGTTFRQTPAPVNNASNMGTYQVMSVESPVS 152
579****************************************************************************************** PP
DUF822 96 .sslkssalaspvesysaspksssfpspssldsislasa.asllpvlsvl 143
ss +++++++ v+ + ++++ ++++sp+s+dsi ++++ +++ +++s+
Sopen08g001780.2 153 gSSFRNCSTRASVDCQPSVTRINESLSPASFDSIVVTESdTKVDKFTSTS 202
**********************************9998864444444443 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 1.6E-50 | 60 | 205 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| SuperFamily | SSF51445 | 6.74E-156 | 229 | 661 | IPR017853 | Glycoside hydrolase superfamily |
| Gene3D | G3DSA:3.20.20.80 | 1.5E-168 | 230 | 661 | IPR013781 | Glycoside hydrolase, catalytic domain |
| Pfam | PF01373 | 2.9E-82 | 254 | 624 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.0E-52 | 269 | 283 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.0E-52 | 290 | 308 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.0E-52 | 312 | 333 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.0E-52 | 405 | 427 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.0E-52 | 478 | 497 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.0E-52 | 512 | 528 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.0E-52 | 529 | 540 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.0E-52 | 547 | 570 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.0E-52 | 583 | 605 | IPR001554 | Glycoside hydrolase, family 14 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000272 | Biological Process | polysaccharide catabolic process | ||||
| GO:0048831 | Biological Process | regulation of shoot system development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0016161 | Molecular Function | beta-amylase activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 667 aa Download sequence Send to blast |
MSNPHHHISN TQDPDPQFPN PNDDPNPNHP LQPQSNPQPR RPRGFAATSA AAAATGTINK 60 SRKEREKEKE RTKLRERHRR AITSRMLAGL RQYGNFPLPV RADMNDVLAA LARQAGWTVE 120 PDGTTFRQTP APVNNASNMG TYQVMSVESP VSGSSFRNCS TRASVDCQPS VTRINESLSP 180 ASFDSIVVTE SDTKVDKFTS TSPMNSTGCL EAGQLMQELH CGEHGGSFSE TQYIPVFVML 240 SQSGVINNFC QLMDPDGVKQ ELQQLKSLKI DGVVVNCWWG IVESWVPQKY EWSGYRELFK 300 IIRDFYMKLQ VVMAFHENGG SDTGGMFISL PQWVLEIGKD NQDIFFTDHQ GRRNTECLSW 360 GIDKERVLRG RTAIEVYFDL MRSFRTEFDD LFADGLISAV EIGLGASGEL KYPSFSERMG 420 WRYPGIGEFQ CYDKYSLQNL RKAATSRGHS FWAKGPDNAG FYNAKPHETG FFCERGDYDS 480 YYGRFFLHWY RQVLIDHADN VLTLASLAFE GIQIVVKIPS IYWWYRTSSH AAELTAGYYN 540 PTNQDGYSPV FEVLKKHSVT IKFVSSGLQV PETDDALADP EGLSWQIMNS AWDKEINVAG 600 QNAFPCYDRE GFMRLVETAK PRNDPDRHRF SFFAFQQPSP LVQSAICFSE LDYFIKCMHG 660 EINNVES |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wdq_A | 1e-121 | 233 | 661 | 11 | 444 | Beta-amylase |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00543 | DAP | Transfer from AT5G45300 | Download |
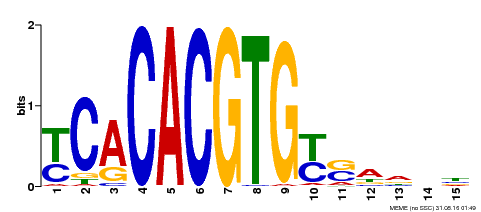
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | HG975447 | 0.0 | HG975447.1 Solanum pennellii chromosome ch08, complete genome. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015085911.1 | 0.0 | beta-amylase 8 isoform X5 | ||||
| Swissprot | Q9FH80 | 0.0 | BAM8_ARATH; Beta-amylase 8 | ||||
| TrEMBL | A0A3Q7IDQ8 | 0.0 | A0A3Q7IDQ8_SOLLC; Beta-amylase | ||||
| STRING | Solyc08g005780.2.1 | 0.0 | (Solanum lycopersicum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA1607 | 24 | 48 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G45300.1 | 0.0 | beta-amylase 2 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



