 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sopen09g001040.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Solaneae; Solanum; Lycopersicon
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 326aa MW: 36402.8 Da PI: 8.3364 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 100.1 | 1.5e-31 | 62 | 115 | 2 | 55 |
G2-like 2 prlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
+rlrWtpeLH rFv+ave+LGG+++AtPk +l++m++kgL+++hvkSHLQ++R+
Sopen09g001040.1 62 ARLRWTPELHLRFVHAVERLGGQDRATPKLVLQMMNIKGLSIAHVKSHLQMFRS 115
69***************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 2.0E-29 | 58 | 116 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 11.958 | 58 | 118 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 4.12E-15 | 61 | 117 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 2.6E-22 | 62 | 117 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 1.1E-9 | 63 | 113 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 326 aa Download sequence Send to blast |
MLEDSGETGC NSKTSCNFEN EVLEENDASS KFKDGGSSSE STLEESEKIK PCVRPYVRSK 60 MARLRWTPEL HLRFVHAVER LGGQDRATPK LVLQMMNIKG LSIAHVKSHL QMFRSKKTDD 120 QSQGIGHHHK LFMEGGDPNI FNMIQFPRFP TYHQRLNSTF RYGDASWNCH GNWMPSNTMG 180 QMIPSFINET STIQRNEIKD VTGSSIGSPS GELTRLFHIA SKAQARAFVG NGITSPPNLE 240 RTITPLKRKV SYSDQVDLNL YLGVKPRNGF LTPNLDDNDN DNGSSLSLSL SSPLSYSRAT 300 RFIEDANIDG KTENARRGAS TLDLTL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 2e-18 | 62 | 117 | 3 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 2e-18 | 62 | 117 | 3 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_A | 2e-18 | 62 | 117 | 2 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 2e-18 | 62 | 117 | 2 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 2e-18 | 62 | 117 | 2 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 2e-18 | 62 | 117 | 2 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 2e-18 | 62 | 117 | 3 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 2e-18 | 62 | 117 | 3 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 2e-18 | 62 | 117 | 3 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 2e-18 | 62 | 117 | 3 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 2e-18 | 62 | 117 | 3 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 2e-18 | 62 | 117 | 3 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00297 | DAP | Transfer from AT2G38300 | Download |
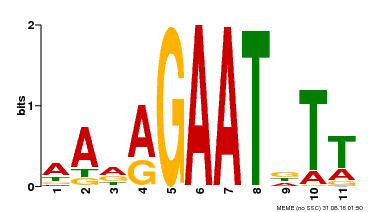
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | HG975448 | 0.0 | HG975448.1 Solanum pennellii chromosome ch09, complete genome. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015087329.1 | 0.0 | putative two-component response regulator ARR21 | ||||
| TrEMBL | M1CJ60 | 0.0 | M1CJ60_SOLTU; Uncharacterized protein | ||||
| STRING | PGSC0003DMT400068508 | 0.0 | (Solanum tuberosum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA4382 | 21 | 41 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G38300.1 | 7e-51 | G2-like family protein | ||||



