 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sopen12g034010.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Solaneae; Solanum; Lycopersicon
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 910aa MW: 103231 Da PI: 7.1063 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 160.8 | 2.6e-50 | 30 | 146 | 3 | 118 |
CG-1 3 ke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptf 94
+e k rwl+++ei+aiL n++ +++ ++ + pksg+++L++rk++r+fr+DGy+wkkkkdgktv+E+he+LKvg+ e +++yYah+e+n+tf
Sopen12g034010.1 30 EEsKMRWLRPNEIHAILCNHKYFNINVKPVNLPKSGTIVLFDRKMLRNFRRDGYNWKKKKDGKTVKEAHEHLKVGNDERIHVYYAHGEDNTTF 122
44499**************************************************************************************** PP
CG-1 95 qrrcywlLeeelekivlvhylevk 118
rrcywlL+++le++vlvhy+e++
Sopen12g034010.1 123 VRRCYWLLDKTLEHVVLVHYRETQ 146
*********************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 74.906 | 25 | 151 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 2.2E-73 | 28 | 146 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 7.3E-45 | 32 | 144 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:2.60.40.10 | 1.5E-5 | 354 | 456 | IPR013783 | Immunoglobulin-like fold |
| SuperFamily | SSF81296 | 1.82E-12 | 367 | 454 | IPR014756 | Immunoglobulin E-set |
| Pfam | PF12796 | 4.1E-7 | 552 | 632 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 5.36E-16 | 552 | 662 | No hit | No description |
| Gene3D | G3DSA:1.25.40.20 | 7.8E-17 | 552 | 666 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 4.04E-18 | 553 | 667 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 15.698 | 561 | 674 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50088 | 11.701 | 603 | 635 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 1.8E-6 | 603 | 632 | IPR002110 | Ankyrin repeat |
| SMART | SM00015 | 14 | 765 | 787 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.236 | 766 | 795 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.1 | 768 | 786 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0021 | 788 | 810 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 10.017 | 789 | 813 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 3.0E-4 | 790 | 810 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 25 | 864 | 886 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.37 | 865 | 894 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 910 aa Download sequence Send to blast |
MESSVSGRLL GCEIHGFRTM QDLDIPNIME ESKMRWLRPN EIHAILCNHK YFNINVKPVN 60 LPKSGTIVLF DRKMLRNFRR DGYNWKKKKD GKTVKEAHEH LKVGNDERIH VYYAHGEDNT 120 TFVRRCYWLL DKTLEHVVLV HYRETQEVSS NSTVAQGSPA APVSSGSALS DPADLSASWV 180 LSGELDSAVD QQYSASRHAH LEPNRDMTVQ NHEQRLLEIN TLEWDDLLAP GDPNKMVATQ 240 QAGGKTAYVQ HTSYEQRNLC ELNGYSLDGG VSSSLERIST FNNSNEITFQ TVDGQMTSSF 300 EKNESGVMTV STGDSLDSLN QDRLQTQDSF GRWMNYLIKD SPESIDDPTP ESSVSTGQSY 360 ASEQIFNITE ILPAWAPSTE ETKICVIGQF HGEQSHLESS SLHCVCGDAC FPAEVLQPGV 420 YRCIVSPQTP GLVNIYLSFD GNKPISQVMS FEFRAPSVHV WTEPPENKSD WDEFRNQMRL 480 AHLLFSTSKS LNILSSKIHQ DLLKDAKKFA GKCSHIIDDW ACLIKSIEDK KVSVPRAKDC 540 LFELSLKTRL QEWLLERVVE GCKISEHDEQ GQGVIHLCAI LGYTWAVYPF SWSGLSLDYR 600 DKYGWTALHW AAYYGREKMV ATLLSAGAKP NLVTDPTSEN LGGCTASDLA SKNGHEGLGA 660 YLAEKALVAQ FKDMTLAGNI SGSLQTTTES INPGNFTEEE LNLKDSLAAY RTAADAAARI 720 QAAFRERALK VRTKAVESSN PEMEARNIIA AMKIQHAFRN YEMQKQLAAA ARIQYRFRTW 780 KMRKEFLHMR RQAIKIQAVF RGFQVRRHYR KIIWSVGVLE KALFRWRLKR KGLRGLKLQS 840 SQVIKPDDVE EDFFQASRKQ AEERIERSVV RVQAMFRSKQ AQEQYRRMKL EHDKATLEYE 900 GTLNPDTEMD |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator (PubMed:14581622). Binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in response to cold. Contributes together with CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:14581622, ECO:0000269|PubMed:28351986, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00435 | DAP | Transfer from AT4G16150 | Download |
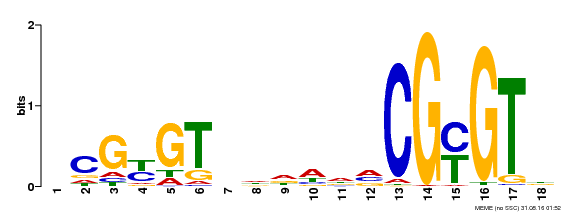
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, wounding, abscisic acid, H(2)O(2) and salicylic acid. {ECO:0000269|PubMed:12218065}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | JN566051 | 0.0 | JN566051.1 Solanum lycopersicum calmodulin-binding transcription factor SR3L mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015059393.1 | 0.0 | calmodulin-binding transcription activator 6 isoform X1 | ||||
| Swissprot | O23463 | 0.0 | CMTA5_ARATH; Calmodulin-binding transcription activator 5 | ||||
| TrEMBL | H8ZRY6 | 0.0 | H8ZRY6_SOLLC; Calmodulin-binding transcription factor SR3L | ||||
| STRING | Solyc12g099340.1.1 | 0.0 | (Solanum lycopersicum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA4565 | 22 | 32 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16150.1 | 0.0 | calmodulin binding;transcription regulators | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



