 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sp_023380_jnpe.t1 | ||||||||
| Common Name | SOVF_023380 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Chenopodiaceae; Chenopodioideae; Anserineae; Spinacia
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 263aa MW: 28418.7 Da PI: 7.1948 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 58 | 2.4e-18 | 37 | 87 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
++ykGVr ++ +g+Wv+eIr p ++++r++lg++ t+e Aa+a++aa ++l+g
Sp_023380_jnpe.t1 37 KKYKGVRMRS-WGSWVSEIRAP---NQKTRIWLGSYSTPEAAARAYDAALLCLKG 87
59*****998.**********9...336************************998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00018 | 5.91E-21 | 37 | 97 | No hit | No description |
| SMART | SM00380 | 1.9E-36 | 38 | 101 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 21.694 | 38 | 95 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 6.54E-20 | 38 | 97 | IPR016177 | DNA-binding domain |
| Gene3D | G3DSA:3.30.730.10 | 4.3E-30 | 38 | 97 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 1.7E-11 | 38 | 87 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.9E-8 | 39 | 50 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.9E-8 | 61 | 77 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0034605 | Biological Process | cellular response to heat | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0071497 | Biological Process | cellular response to freezing | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 263 aa Download sequence Send to blast |
MVKSELKKTE SEAAKPMSSS SSSLSSSLSL SSSSLKKKYK GVRMRSWGSW VSEIRAPNQK 60 TRIWLGSYST PEAAARAYDA ALLCLKGSSA NLNFPHSSYS TTCPLIPSSN NHTTTTTTTT 120 TTVTTVMSPK SIQRVAAAAA AAAATTSTTT TFTDQFHHCS SSSSISPSPS MSSSPSLSSS 180 SPPLGQITDD LISLTHPYPS FNADINNALM MNNNEEDPWS WFDVSPKHVD LMLNSMFFNP 240 PSLNIMDIEQ LHDDGDIPLW SFR |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5wx9_A | 4e-15 | 38 | 97 | 14 | 74 | Ethylene-responsive transcription factor ERF096 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00156 | DAP | Transfer from AT1G21910 | Download |
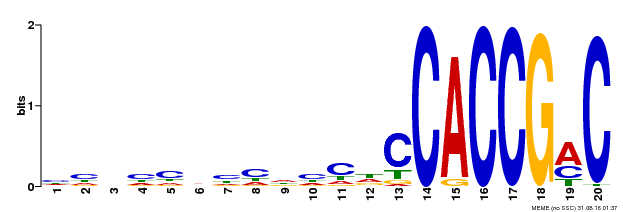
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021855735.1 | 0.0 | ethylene-responsive transcription factor ERF014-like | ||||
| TrEMBL | A0A0K9RXR2 | 0.0 | A0A0K9RXR2_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010670581.1 | 2e-57 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G21910.1 | 5e-32 | ERF family protein | ||||



