 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sp_042210_agke.t1 | ||||||||
| Common Name | SOVF_042210 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Chenopodiaceae; Chenopodioideae; Anserineae; Spinacia
|
||||||||
| Family | GATA | ||||||||
| Protein Properties | Length: 283aa MW: 31386.9 Da PI: 9.6776 | ||||||||
| Description | GATA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GATA | 54.4 | 1.8e-17 | 204 | 237 | 1 | 34 |
GATA 1 CsnCgttkTplWRrgpdgnktLCnaCGlyyrkkg 34
C +Cg+ kTp+WR gp+g+ktLCnaCG++y++ +
Sp_042210_agke.t1 204 CLHCGSEKTPQWRAGPSGPKTLCNACGVRYKSGR 237
99*****************************987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF57716 | 4.13E-14 | 197 | 259 | No hit | No description |
| PROSITE profile | PS50114 | 12.716 | 198 | 234 | IPR000679 | Zinc finger, GATA-type |
| SMART | SM00401 | 4.6E-15 | 198 | 252 | IPR000679 | Zinc finger, GATA-type |
| Gene3D | G3DSA:3.30.50.10 | 3.4E-14 | 199 | 235 | IPR013088 | Zinc finger, NHR/GATA-type |
| CDD | cd00202 | 7.90E-15 | 203 | 251 | No hit | No description |
| PROSITE pattern | PS00344 | 0 | 204 | 229 | IPR000679 | Zinc finger, GATA-type |
| Pfam | PF00320 | 3.8E-15 | 204 | 238 | IPR000679 | Zinc finger, GATA-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0007623 | Biological Process | circadian rhythm | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 283 aa Download sequence Send to blast |
MESFDKAARF MDELLDFSSD IGEEESDDEI PGNSSKSHHF LHHHRLNHRR HNRQSPPFAE 60 FVEEELEWIS NKDAFPMVES LVDILPNHPR IASSYNTNHQ SPISVLDNSN SSSSSSANSH 120 LTHGSLNNNN INSNGGRGGN MMLINLSSDF KVPGKARSKH GRKRRKDMLG SQFKCFVYEK 180 KGARKLASSV SASSKSSSTL GRKCLHCGSE KTPQWRAGPS GPKTLCNACG VRYKSGRLCD 240 EYRPASSPTF SSDLHSNSHR KIVEMRRQKM PLTGSGSNSL EIG |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 161 | 166 | RKRRKD |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00377 | DAP | Transfer from AT3G24050 | Download |
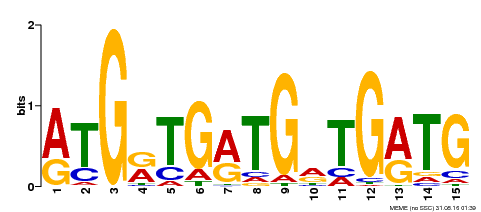
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021839064.1 | 0.0 | GATA transcription factor 1-like isoform X2 | ||||
| TrEMBL | A0A0K9RPU0 | 0.0 | A0A0K9RPU0_SPIOL; Uncharacterized protein | ||||
| STRING | EOY05429 | 5e-69 | (Theobroma cacao) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G24050.1 | 5e-47 | GATA transcription factor 1 | ||||



