 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sp_049570_zwtw.t1 | ||||||||
| Common Name | SOVF_049570 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Chenopodiaceae; Chenopodioideae; Anserineae; Spinacia
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 649aa MW: 69382.9 Da PI: 9.5161 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 16 | 3.4e-05 | 122 | 144 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
+ C+ C+k F r nL+ H r H
Sp_049570_zwtw.t1 122 FICEVCNKGFQREQNLQLHRRGH 144
89*******************88 PP
| |||||||
| 2 | zf-C2H2 | 13.7 | 0.00019 | 199 | 221 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
+kC++C+k++ +s+ k H +t+
Sp_049570_zwtw.t1 199 FKCEKCSKRYAVQSDWKAHSKTC 221
89*****************9998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF81995 | 8.11E-5 | 23 | 122 | No hit | No description |
| SuperFamily | SSF57667 | 6.15E-8 | 121 | 144 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 7.4E-6 | 121 | 144 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| Pfam | PF12171 | 3.2E-5 | 122 | 144 | IPR022755 | Zinc finger, double-stranded RNA binding |
| SMART | SM00355 | 0.0052 | 122 | 144 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 10.99 | 122 | 144 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 124 | 144 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 150 | 164 | 194 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 1.2E-5 | 187 | 220 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 6.15E-8 | 194 | 219 | No hit | No description |
| SMART | SM00355 | 88 | 199 | 219 | IPR015880 | Zinc finger, C2H2-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 649 aa Download sequence Send to blast |
MAASSSSSAF FGSRDGDAMK HGHHQLQLQH QPPQQPSQQV LQPQQQQQLQ QQQQQQQQQQ 60 QQQQQQQQQH SSSVPQPPSS SSAPPSNPQQ KKKRSLPGTP SKYPDAEVIA LSPKTLMATN 120 RFICEVCNKG FQREQNLQLH RRGHNLPWKL RQKTNKDQVR KKVYLCPEPT CVHHDSARAL 180 GDLTGIKKHY SRKHGEKKFK CEKCSKRYAV QSDWKAHSKT CGTREYRCDC GTLFSRRDSF 240 ITHRAFCDAL AQESARHPTS LSTLGSHLFG PSSALGGANN GANNNMGLGV GLSHQVANHL 300 PPSLSDQMTS DILRLGGGAR GVGSSQFDHL LGPGSFRQQH NIQQNQANFY LSDQTGGSNH 360 FANSNNNDQE NQSQNAGGGI FSNKLFTGQL PVPDLGFFSN NNNNENTNTI NTNTNAGSGD 420 HNVLFSPTGS GSLFGGPPSG GLSSLYTTGP ALQGSPHMSA TALLQKAAQM GSTTSTSNAS 480 ATALLKGLEA KQTPQQQSAP PQPPTSVGGG GFGGLFGEND QNNLHDLLMR DNLMSSPFFG 540 GGDHHHHQTG GTGDHNNNNY GGFNPNKTGL GVGSSDRLTR DFLGVGEIVR SMSGSGGGFP 600 PNSQSMASLD LPADHQRNNS AAAAAATNSN NVNNSAPSSS QAFGGGNTF |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5b3h_C | 2e-33 | 195 | 256 | 3 | 64 | Zinc finger protein JACKDAW |
| 5b3h_F | 2e-33 | 195 | 256 | 3 | 64 | Zinc finger protein JACKDAW |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 148 | 161 | KLRQKTNKDQVRKK |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00255 | DAP | Transfer from AT2G02070 | Download |
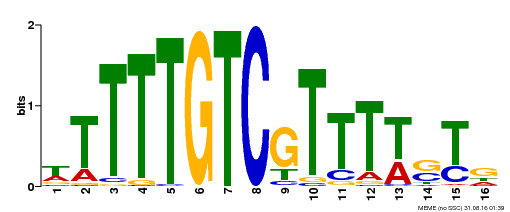
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021856017.1 | 0.0 | protein indeterminate-domain 9-like isoform X1 | ||||
| TrEMBL | A0A0K9RP19 | 0.0 | A0A0K9RP19_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010680044.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G02070.1 | 1e-107 | indeterminate(ID)-domain 5 | ||||



