 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sp_057210_wxdz.t1 | ||||||||
| Common Name | SOVF_057210 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Chenopodiaceae; Chenopodioideae; Anserineae; Spinacia
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 376aa MW: 41409.8 Da PI: 4.8131 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 61.6 | 1.8e-19 | 147 | 197 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkr.krfslgkfgtaeeAakaaiaarkkleg 55
++++GVr+++ +g+W+AeIrdp r +r +lg+++taeeAa +++a+ +l+g
Sp_057210_wxdz.t1 147 KKFRGVRQRP-WGKWAAEIRDPL----RrQRLWLGTYDTAEEAAMVYDNAAIQLRG 197
59********.**********73....45*************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00018 | 1.29E-32 | 147 | 204 | No hit | No description |
| SuperFamily | SSF54171 | 7.19E-22 | 148 | 206 | IPR016177 | DNA-binding domain |
| Gene3D | G3DSA:3.30.730.10 | 2.3E-30 | 148 | 205 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 9.0E-13 | 148 | 197 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 3.9E-38 | 148 | 211 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 23.578 | 148 | 205 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 4.7E-10 | 149 | 160 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 4.7E-10 | 171 | 187 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0042991 | Biological Process | transcription factor import into nucleus | ||||
| GO:0048364 | Biological Process | root development | ||||
| GO:0048825 | Biological Process | cotyledon development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 376 aa Download sequence Send to blast |
MMESPQTFPP IKYTEHRNQT ILQKQIRNKS YPDINNHSVY PKIVRISVTD ADATDSSSDE 60 EYDVAASDDF RRRRIKRFIN EVSIEPCSSS SYVSAPTVSS SEGLRESGGA VAEVAANGAV 120 SKRPKKRGKC SSVKQVQGTN GNGNGGKKFR GVRQRPWGKW AAEIRDPLRR QRLWLGTYDT 180 AEEAAMVYDN AAIQLRGPNA LTNFSTPHNP DNHQQQPPQE LKPKKLSLST TSSCVDDDED 240 GSHLAPSICS PTSVLRFQPE PESEPISPKE EAQVVKEEEN VQPLGPMVSS PYVSCVDSLF 300 PSGFFDFDTT VPSVPDLFEQ PGFGSDGGHV FIDDCKLGMF FDHHDDGDFL GLSGWSSNDY 360 FPDIGDIFGS DPLVAI |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2gcc_A | 7e-21 | 146 | 204 | 3 | 62 | ATERF1 |
| 3gcc_A | 7e-21 | 146 | 204 | 3 | 62 | ATERF1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00027 | PBM | Transfer from AT4G23750 | Download |
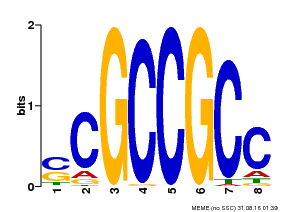
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021838632.1 | 0.0 | ethylene-responsive transcription factor CRF2-like | ||||
| TrEMBL | A0A0K9RK75 | 0.0 | A0A0K9RK75_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010676352.1 | 1e-116 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G23750.2 | 3e-44 | cytokinin response factor 2 | ||||



