 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sp_123000_huqx.t1 | ||||||||
| Common Name | SOVF_123000 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Chenopodiaceae; Chenopodioideae; Anserineae; Spinacia
|
||||||||
| Family | GRAS | ||||||||
| Protein Properties | Length: 615aa MW: 66534.2 Da PI: 5.2593 | ||||||||
| Description | GRAS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GRAS | 442.5 | 3e-135 | 246 | 604 | 1 | 374 |
GRAS 1 lvelLlecAeavssgdlelaqalLarlselaspdgdpmqRlaayfteALaarlarsvselykalppsetseknsseelaalklfsevsPilk 92
lv++L++cAea++++++ la+alL+++ la+++ +m+++a+yf+eALa+r+++ + p + n s+ l++ f+e++P+lk
Sp_123000_huqx.t1 246 LVHTLMACAEAIEQQNMGLAEALLKQIGFLAASQVGSMRKVATYFAEALARRVYK--------VCPDVPYDGNLSDMLQM--HFYETCPYLK 327
689***************************************************9........22222333446666555..5********* PP
GRAS 93 fshltaNqaIleavegeervHiiDfdisqGlQWpaLlqaLasRpegppslRiTgvgspesgskeeleetgerLakfAeelgvpfefnvlvak 184
f+h+taNqaIlea++g+++vH+iDf++++G+QWpaL+qaLa Rpegpp +R+Tg+g+p+++++++l+e+g++La+fA++++++fe++ +va+
Sp_123000_huqx.t1 328 FAHFTANQAILEAFSGKKKVHVIDFSMKEGMQWPALMQALALRPEGPPAFRLTGIGPPAPDNSDRLQEVGWKLAQFADSIQIQFEYRGFVAN 419
******************************************************************************************** PP
GRAS 185 rledleleeLrvkp.gEalaVnlvlqlhrlldesvsleserdevLklvkslsPkvvvvveqeadhnsesFlerflealeyysalfdsleakl 275
+l+dle L+++p E++aVn+v++lhrll+++++ ++ vL l+k+++P +v+vveqea+hn++ Flerf e+l+yys+lfdsle+ +
Sp_123000_huqx.t1 420 SLTDLESALLELRPdTEVVAVNSVFELHRLLARPGAADK----VLGLMKEVNPVIVTVVEQEANHNGPAFLERFNESLHYYSTLFDSLESCV 507
**************99***************99988888....************************************************7 PP
GRAS 276 preseerikvErellgreivnvvacegaerrerhetlekWrerleeaGFkpvplsekaakqaklllrkvk.sdgyrveeesgslvlgWkdrp 366
+++++++++++lg +i+nvvaceg +r+erhet+++Wr r+++aGF pv+++++a kqa++ll ++ +dgy vee+ g+l+lgW++rp
Sp_123000_huqx.t1 508 ---DSQDKMMSEMYLGSQICNVVACEGVDRVERHETVAQWRTRFGKAGFAPVHIGSNAFKQASMLLDFFAgGDGYGVEENGGCLMLGWHSRP 596
...88889999999****************************************************************************** PP
GRAS 367 LvsvSaWr 374
L+++SaW+
Sp_123000_huqx.t1 597 LITTSAWQ 604
*******6 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01129 | 1.9E-37 | 43 | 116 | No hit | No description |
| Pfam | PF12041 | 7.6E-37 | 43 | 109 | IPR021914 | Transcriptional factor DELLA, N-terminal |
| PROSITE profile | PS50985 | 65.501 | 220 | 583 | IPR005202 | Transcription factor GRAS |
| Pfam | PF03514 | 1.0E-132 | 246 | 604 | IPR005202 | Transcription factor GRAS |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009867 | Biological Process | jasmonic acid mediated signaling pathway | ||||
| GO:0009938 | Biological Process | negative regulation of gibberellic acid mediated signaling pathway | ||||
| GO:0010187 | Biological Process | negative regulation of seed germination | ||||
| GO:0010218 | Biological Process | response to far red light | ||||
| GO:0042176 | Biological Process | regulation of protein catabolic process | ||||
| GO:0042538 | Biological Process | hyperosmotic salinity response | ||||
| GO:2000033 | Biological Process | regulation of seed dormancy process | ||||
| GO:2000377 | Biological Process | regulation of reactive oxygen species metabolic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 615 aa Download sequence Send to blast |
MKRELPFSNP GPSAGGGGSL GGGPPTTTGK SKMWEDQNDA GVDELLAVLG YKVRSSDMAE 60 VAQKLEQLEQ AMGNVREDGL SQLASETVHY NPSDLSTWLE SMLSEFNPSS TSFDPANPIL 120 LPSIGSRVDP IIVNPTPQGS VKFGGSDPYS DYDLKAIPGK AILTPPSPSS SALVVNNSSS 180 SNSLVSTTTS SSSSREAKRL KASSYTNPQS AKATASGLTS SILTTNTAVS SRPVVLVDSQ 240 ENGVRLVHTL MACAEAIEQQ NMGLAEALLK QIGFLAASQV GSMRKVATYF AEALARRVYK 300 VCPDVPYDGN LSDMLQMHFY ETCPYLKFAH FTANQAILEA FSGKKKVHVI DFSMKEGMQW 360 PALMQALALR PEGPPAFRLT GIGPPAPDNS DRLQEVGWKL AQFADSIQIQ FEYRGFVANS 420 LTDLESALLE LRPDTEVVAV NSVFELHRLL ARPGAADKVL GLMKEVNPVI VTVVEQEANH 480 NGPAFLERFN ESLHYYSTLF DSLESCVDSQ DKMMSEMYLG SQICNVVACE GVDRVERHET 540 VAQWRTRFGK AGFAPVHIGS NAFKQASMLL DFFAGGDGYG VEENGGCLML GWHSRPLITT 600 SAWQLAQNSK SPIRH |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5b3g_A | 7e-61 | 241 | 603 | 14 | 378 | Protein SCARECROW |
| 5b3h_A | 6e-61 | 241 | 603 | 13 | 377 | Protein SCARECROW |
| 5b3h_D | 6e-61 | 241 | 603 | 13 | 377 | Protein SCARECROW |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcriptional regulator that acts as a repressor of the gibberellin (GA) signaling pathway. Probably acts by participating in large multiprotein complexes that repress transcription of GA-inducible genes. Upon GA application, it is degraded by the proteasome, allowing the GA signaling pathway. | |||||
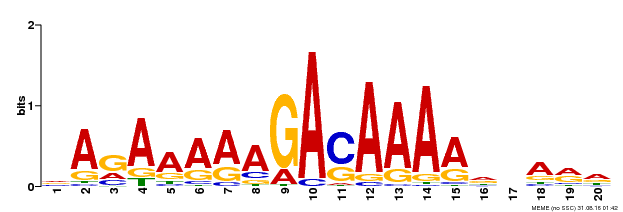
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00611 | ChIP-seq | Transfer from AT2G01570 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021864610.1 | 0.0 | DELLA protein GAIP-like | ||||
| Swissprot | Q8S4W7 | 0.0 | GAI1_VITVI; DELLA protein GAI1 | ||||
| TrEMBL | A0A0K9R1U0 | 0.0 | A0A0K9R1U0_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010681882.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G01570.1 | 0.0 | GRAS family protein | ||||



