 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sp_123350_gdgu.t1 | ||||||||
| Common Name | SOVF_123350 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Chenopodiaceae; Chenopodioideae; Anserineae; Spinacia
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 1078aa MW: 118370 Da PI: 7.9207 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 126.9 | 7.7e-40 | 163 | 240 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+Cqv++C++dls+ k+yhrrhkvCe+hsk++++lv +++qrfCqqCsrfh l efDe+krsCrrrLa+hn+rrrk+qa
Sp_123350_gdgu.t1 163 MCQVDNCTEDLSKSKDYHRRHKVCEFHSKSTKALVGKQMQRFCQQCSRFHPLAEFDEGKRSCRRRLAGHNKRRRKTQA 240
6**************************************************************************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 4.6E-33 | 157 | 225 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 31.385 | 161 | 238 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 1.57E-36 | 162 | 241 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 3.4E-29 | 164 | 237 | IPR004333 | Transcription factor, SBP-box |
| CDD | cd00204 | 1.26E-7 | 826 | 963 | No hit | No description |
| SuperFamily | SSF48403 | 1.28E-8 | 828 | 968 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 1.5E-7 | 832 | 971 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 340 | 867 | 896 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 3.2 | 912 | 942 | IPR002110 | Ankyrin repeat |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005886 | Cellular Component | plasma membrane | ||||
| GO:0016021 | Cellular Component | integral component of membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1078 aa Download sequence Send to blast |
MEEVGTQVAS PLFIHQNIGG RFCEGALIGA KRGLCYNSGT NHHQQQQHSQ NHRSGHGWNP 60 KDWEWDSSHF VARNKPIEAS QRVNPILSSS FTSKSSSRVD DQQEQDGGGS LRLQLGGVGD 120 SNVTGTSINK VQSSNSTDDP ASSSRPNKKV RSGSPGTGGN HPMCQVDNCT EDLSKSKDYH 180 RRHKVCEFHS KSTKALVGKQ MQRFCQQCSR FHPLAEFDEG KRSCRRRLAG HNKRRRKTQA 240 EDTPSPPIQP TKTAYGNLDI VNLLTVLARG QGNAEQNVST CPSLPDKNQL IQILSKMNTL 300 PMPLDIATNP LVTPGSTKNG FEQGALMEVN AASRSTTDLL AVLSSTLAAS SPNSVPFISQ 360 RSNPGSCMVK NKSTTGDKDT GSDACKKTAI ELQSLGGERS SSSYQSPTED SDSQVQDTRM 420 NLPLQLFSSS PGDDSSPNLV SSRRYFSSDS SNPTEERSHS STAPVTRKLF PLETTSGSAK 480 PARMSFGEEA NVNIEATSGS RMTLDLFTMG NRAVSNSLQS LPHQAGYTSS SGSDHSPPSF 540 NSDPQKDRTG RIIFKLFDKD PSQLPGALRT QIYNWLSNSP TEMESYIRPG CVVLSIYVSM 600 SSAAWEQLEE SFLQRVEALV QDPDFEFWRS GRFSANIGRQ LAVHKDGRIR ICKPWSTVSS 660 PELFLVSPLA VVSGQSTSLV LGGRNLTSPG TKIHCTYMGG YSSKAVLRSS DEGFTCDEVE 720 LSDFKVHAAA SSVLGRCFIE VENGVRGNCF PVIIADAKIC QELRLLEREF YEAKVGDVVM 780 EDQVQYVASP HSHEEALHFL NELGWLFQRQ LSSDIAVHDF MLHRLQYLLT FSTERDYCAL 840 VKTLLDIFVE AESRMDGLSA ECVEALAKMH LLHRAVKRSS KKMVDMLIHY SAPCGSGASK 900 KYIFPPNLRG PGGITPLHLA ACTSGSYDII DVLTDDPMQI GLHCWKSLVD DSGQSPYSYA 960 LMRNNNAVNN IVARKLSDRI NRQVSVTIGN EIVESFSGTA EMKQRASLKL NTKQNSCSKC 1020 AMMSYGRMQG SHGFLHRPFI HSMLTIAAVC VCVCLFFKSM HVNSVTPFMW DNVDFGAI |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 3e-30 | 153 | 237 | 1 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3' (By similarity). May play a role in plant development. {ECO:0000250, ECO:0000269|PubMed:15703061}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00155 | DAP | Transfer from AT1G20980 | Download |
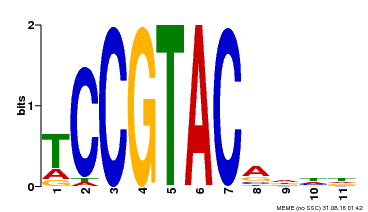
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021836587.1 | 0.0 | squamosa promoter-binding-like protein 14 | ||||
| Refseq | XP_021836588.1 | 0.0 | squamosa promoter-binding-like protein 14 | ||||
| Swissprot | Q8RY95 | 0.0 | SPL14_ARATH; Squamosa promoter-binding-like protein 14 | ||||
| TrEMBL | A0A0K9R1B9 | 0.0 | A0A0K9R1B9_SPIOL; Uncharacterized protein | ||||
| STRING | VIT_18s0122g00380.t01 | 0.0 | (Vitis vinifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G20980.1 | 0.0 | squamosa promoter binding protein-like 14 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



