 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sp_126330_ypqg.t1 | ||||||||
| Common Name | SOVF_126330 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Chenopodiaceae; Chenopodioideae; Anserineae; Spinacia
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 1006aa MW: 112305 Da PI: 6.0213 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 184.4 | 1.2e-57 | 22 | 137 | 3 | 118 |
CG-1 3 kekkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptf 94
+ ++rwl++ ei++iL nf+k+++++e++trp+sgsl+L++rk++ryfrkDG++w+kkkdgktv+E+hekLKvg+v+vl+cyYah+e+n f
Sp_126330_ypqg.t1 22 EAQHRWLRPAEICEILRNFQKFQISSEPPTRPSSGSLFLFDRKVLRYFRKDGHNWRKKKDGKTVKEAHEKLKVGSVDVLHCYYAHGEDNDCF 113
349***************************************************************************************** PP
CG-1 95 qrrcywlLeeelekivlvhylevk 118
qrr+ywlLeee+ +iv+vhylevk
Sp_126330_ypqg.t1 114 QRRSYWLLEEEFMHIVFVHYLEVK 137
*********************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 84.371 | 16 | 142 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 1.5E-82 | 19 | 137 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 9.1E-51 | 22 | 136 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:2.60.40.10 | 2.6E-7 | 429 | 516 | IPR013783 | Immunoglobulin-like fold |
| SuperFamily | SSF81296 | 7.84E-17 | 431 | 516 | IPR014756 | Immunoglobulin E-set |
| Pfam | PF01833 | 1.3E-5 | 431 | 516 | IPR002909 | IPT domain |
| Gene3D | G3DSA:1.25.40.20 | 4.0E-16 | 589 | 734 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 6.84E-15 | 598 | 708 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 14.504 | 603 | 716 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 2.96E-8 | 633 | 704 | No hit | No description |
| SuperFamily | SSF52540 | 9.82E-8 | 818 | 870 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 0.16 | 820 | 842 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.712 | 821 | 850 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.0014 | 822 | 841 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.006 | 843 | 865 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.377 | 844 | 868 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 3.6E-4 | 847 | 865 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0050826 | Biological Process | response to freezing | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1006 aa Download sequence Send to blast |
MADRGSMNTP GIRLDIQQLQ LEAQHRWLRP AEICEILRNF QKFQISSEPP TRPSSGSLFL 60 FDRKVLRYFR KDGHNWRKKK DGKTVKEAHE KLKVGSVDVL HCYYAHGEDN DCFQRRSYWL 120 LEEEFMHIVF VHYLEVKGNR SSMRDTEPVV SSPQFSNSVS SSISNSYNEA FTGNAGSPSA 180 VSSLTSSYED IESEDNHQAM SKYPSPLNFP LVDDSFENNK LGSLMNSNFL PTYPDDYKGG 240 RLSGPGLDYV SLVQRGAGTN DYNQLRTLDL GSWEEVFQQC TNDSGNLPHN PLISGAPLVG 300 STNGFNESFT QLLDSGFGVN SASAGSLAPF LQNNNPLEFS STEFQDNTNI STTAKQPLLG 360 NIRAEEGLQK VDSFNKWMTK ELAGVDDLDL KSSSNLSWQN IESASAVDDP SIQLENYSMS 420 PSIGQDQLFD IQDFSPSCAS TDSETKVVIT GKFLVSPSDV CKYKWSCMFG EVEVPAEVLG 480 NGVLCCYAPL HSVGQVPFYV TCSNRFACSQ VREFEYLVEG QKANIINGSG NSMTEKQLWL 540 RLQKLLSLSS HGNFDSTSEN IRKKEPIFRK IFLLMEDELC LGVELHLKEK VVSWLFDKVS 600 DDGKGPNMLD EEGQGLLHLA AALAFDWIIP PTLAAGVSVN FRDINGWTAL HWAAFYGREK 660 TVAFLVALEA ASGALTDPSP EFPLGRTAAD LASANGYKGI SGFLAEHSLT AHLETLTMTD 720 RKKDSPVESS MSKAVQTMRE KVVTPGDEGD GSDLSLKDSL AAVRNATQAA GRIHQVFRMQ 780 SFQRKQVSKG VGGDESLLSD EQFLSLVSSK IKRPGRPDEQ SHSAATHIQK KFRGWKKRKE 840 FLIIRERVVR IQAHVRGHQV RKRYKPVVWS VGILEKAILR WRRKGQGLRG FRSDALNKAS 900 STQGTQVVQK KPSEEDDYDY LKEGRKQTEE RLQKALVRVK SMVQYPEGRA QYRRLLTAVE 960 GFQKNQQPYD NSTLNSSENV PAEGEDDMID VDSLLDDDNF MAIAFE |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 909 | 925 | KKPSEEDDYDYLKEGRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds calmodulin in a calcium-dependent manner in vitro (PubMed:11925432). Binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Involved in freezing tolerance (PubMed:19270186). Involved in freezing tolerance in association with CAMTA2 and CAMTA3. Contributes together with CAMTA2 and CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:23581962). Involved in drought stress responses by regulating several drought-responsive genes (PubMed:23547968). Involved in auxin signaling and responses to abiotic stresses (PubMed:20383645). Activates the expression of the V-PPase proton pump AVP1 in pollen (PubMed:14581622). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:11925432, ECO:0000269|PubMed:14581622, ECO:0000269|PubMed:19270186, ECO:0000269|PubMed:20383645, ECO:0000269|PubMed:23547968, ECO:0000269|PubMed:23581962, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00501 | DAP | Transfer from AT5G09410 | Download |
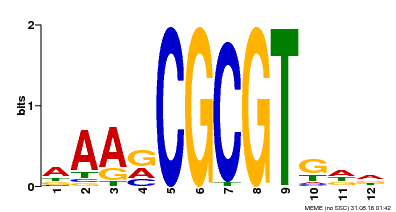
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by UVB, wounding, ethylene and methyl jasmonate (PubMed:12218065). Induced by salt stress and heat shock (PubMed:12218065, PubMed:20383645). Induced by aluminum (PubMed:25627216). {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:20383645, ECO:0000269|PubMed:25627216}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021860116.1 | 0.0 | calmodulin-binding transcription activator 2-like | ||||
| Swissprot | Q9FY74 | 0.0 | CMTA1_ARATH; Calmodulin-binding transcription activator 1 | ||||
| TrEMBL | A0A0K9QYQ2 | 0.0 | A0A0K9QYQ2_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010690491.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G09410.2 | 0.0 | ethylene induced calmodulin binding protein | ||||



