 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Sp_144720_excs.t1 | ||||||||
| Common Name | SOVF_144720 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Chenopodiaceae; Chenopodioideae; Anserineae; Spinacia
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 704aa MW: 77676 Da PI: 4.9036 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 40.7 | 5.6e-13 | 651 | 700 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHT...TTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmg...kgRtlkqcksrwqkyl 48
r++W+ eE++ l avk+lG + Wk+I ++ g +gR+ ++k++w++ l
Sp_144720_excs.t1 651 RKKWSVEEEDTLRVAVKELGRN-WKLILQCHGdafEGRSNVDLKDKWRNML 700
789*****************66.*************************975 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 13.596 | 648 | 704 | IPR017930 | Myb domain |
| SMART | SM00717 | 5.5E-10 | 650 | 702 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.3E-12 | 651 | 704 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 3.5E-10 | 651 | 700 | IPR001005 | SANT/Myb domain |
| CDD | cd11660 | 8.42E-13 | 652 | 701 | No hit | No description |
| SuperFamily | SSF46689 | 3.18E-12 | 653 | 702 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009901 | Biological Process | anther dehiscence | ||||
| GO:0010152 | Biological Process | pollen maturation | ||||
| GO:0043067 | Biological Process | regulation of programmed cell death | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 704 aa Download sequence Send to blast |
MDEKVAKWII EFLLRKPIDE KIINGVLSCL PLSNNDKRLK KTILLRKIES AISDGSVSEK 60 LLELLEMVDE LNHGAGKTVP DSMKRAYCAV AVDCTVRFLE ENVEENGKYF EAVERVWRDR 120 VCKVESLVSE ELNRWLEDIE AAVWDSSVCE KILMRNTRNE ALRAVKRYLK EAWESMGPSF 180 LELVALKVGN DVGELCRSEG GGGGDGGGGG GSGSSGGGGS GGVVGSDNPC RTLIDPQVKT 240 RRPRGIKIFD AVEAAETSMN TANDIQTREM DKVQETLQPS STQQHGAAND SHPSTSRTAH 300 NLSCGKNNDA PIYKMDKVQE ALQPSSTQLH GAVNDSHPST SRTARNLSCG KDNDAPIFKM 360 DKVQEALESN SFHLHADVSD SHLDPLEAVH NMSVTKVTDN MSNLNAIKKQ TTHETPEVRR 420 VQEALNFSAR ELHEAVTDPL PEALEIAKTL SSAVGATLNH GTGHVTGVEN QNTSNDDVLE 480 NQTTSNDDVL ENQNTGNDGV FENHNTGSDT VFENHNTGND AVLENQNTGN DAVLENRNTG 540 NDDVLENQIQ PPVIHATEKN TENVEEEMPE LAGDAQGNNE TDDPKKQHRS KRSLMEANGT 600 SHTFEWDDSI DNVNRGSELH LPPSPKRSKV TPLEQYEDKP KPKPQPLIKG RKKWSVEEED 660 TLRVAVKELG RNWKLILQCH GDAFEGRSNV DLKDKWRNML RSQL |
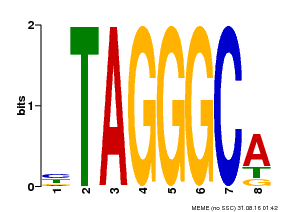
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00052 | PBM | Transfer from AT1G15720 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021857415.1 | 0.0 | uncharacterized protein LOC110796643 isoform X3 | ||||
| TrEMBL | A0A0K9QV37 | 0.0 | A0A0K9QV37_SPIOL; Uncharacterized protein | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G15720.1 | 2e-17 | TRF-like 5 | ||||



